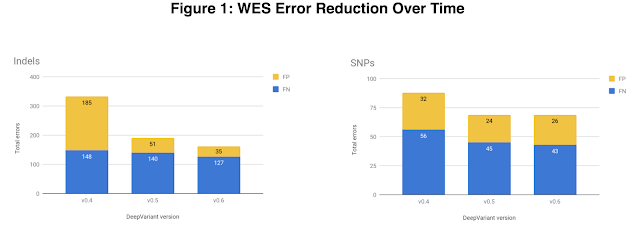
Eric Christiansen, Senior Software Engineer, Google ResearchIn the fields of biology and medicine,
microscopy allows researchers to observe details of cells and molecules which are unavailable to the naked eye.
Transmitted light microscopy, where a biological sample is illuminated on one side and imaged, is relatively simple and well-tolerated by living cultures but produces images which can be difficult to properly assess.
Fluorescence microscopy, in which biological objects of interest (such as cell nuclei) are specifically targeted with fluorescent molecules, simplifies analysis but requires complex sample preparation. With the increasing application of machine learning to the field of microscopy, including algorithms used to
automatically assess the quality of images and
assist pathologists diagnosing cancerous tissue, we wondered if we could develop a deep learning system that could combine the benefits of both microscopy techniques while minimizing the downsides.
With “
In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images”, appearing today in
Cell, we show that a deep neural network can predict fluorescence images from transmitted light images, generating labeled, useful, images without modifying cells and potentially enabling longitudinal studies in unmodified cells, minimally invasive cell screening for cell therapies, and investigations using large numbers of simultaneous labels. We also
open sourced our network, along with the complete training and test data, a trained model checkpoint, and example code.
BackgroundTransmitted light microscopy techniques are easy to use, but can produce images in which it can be hard to tell what’s going on. An example is the following image from a
phase-contrast microscope, in which the intensity of a pixel indicates the degree to which light was
phase-shifted as it passed through the sample.
 |
| Transmitted light (phase-contrast) image of a human motor neuron culture derived from induced pluripotent stem cells. Outset 1 shows a cluster of cells, possibly neurons. Outset 2 shows a flaw in the image obscuring underlying cells. Outset 3 shows neurites. Outset 4 shows what appear to be dead cells. Scale bar is 40 μm. Source images for this and the following figures come from the Finkbeiner lab at the Gladstone Institutes. |
In the above figure, it’s difficult to tell how many cells are in the cluster in Outset 1, or the locations and states of the cells in Outset 4 (hint: there’s a barely-visible flat cell in the upper-middle). It’s also difficult to get fine structures consistently in focus, such as the neurites in Outset 3.
We can get more information out of transmitted light microscopy by acquiring images in z-stacks: sets of images registered in (
x, y) where
z (the distance from the camera) is systematically varied. This causes different parts of the cells to come in and out of focus, which provides information about a sample’s 3D structure. Unfortunately, it often takes a trained eye to make sense of the z-stack, and analysis of such z-stacks has largely defied automation. An example z-stack is shown below.
 |
| A phase-contrast z-stack of the same cells. Note how the appearance changes as the focus is shifted. Now we can see that the fuzzy shape in the lower right of Outset 1 is a single oblong cell, and that the rightmost cell in Outset 4 is taller than the uppermost cell, possibly indicating that it has undergone programmed cell death. |
In contrast, fluorescence microscopy images are easier to analyze, because samples are prepared with carefully engineered fluorescent labels which light up just what the researchers want to see. For example, most human cells have exactly one nucleus, so a nuclear label (such as the blue one below) makes it possible for simple tools to find and count cells in an image.
 |
| Fluorescence microscopy image of the same cells. The blue fluorescent label localizes to DNA, highlighting cell nuclei. The green fluorescent label localizes to a protein found only in dendrites, a neural substructure. The red fluorescent label localizes to a protein found only in axons, another neural substructure. With these labels it is much easier to understand what’s happening in the sample. For example, the green and red labels in Outset 1 confirm this is a neural cluster. The red label in Outset 3 shows that the neurites are axons, not dendrites. The upper-left blue dot in Outset 4 reveals a previously hard-to-see nucleus, and the lack of a blue dot for the cell at the left shows it to be DNA-free cellular debris. |
However, fluorescence microscopy can have significant downsides. First, there is the complexity and variability introduced by the sample preparation and fluorescent labels themselves. Second, when there are many different fluorescent labels in a sample, spectral overlap can make it hard to tell which color belongs to which label, typically limiting researchers to three or four simultaneous labels in a sample. Third, fluorescence labeling may be toxic to cells and sometimes involves protocols that outright kill them, which makes labeling difficult to use in longitudinal studies where the same cells are followed through time.
Seeing more with deep learningIn our paper, we show that a deep neural network can predict fluorescence images from transmitted light z-stacks. To do this, we created a dataset of transmitted light z-stacks matched to fluorescence images and trained a neural network to predict the fluorescence images from the z-stacks. The following diagram explains the process.
 |
| Overview of our system. (A) The dataset of training examples: pairs of transmitted light images from z-stacks with pixel-registered sets of fluorescence images of the same scene. Several different fluorescent labels were used to generate fluorescence images and were varied between training examples; the checkerboard images indicate fluorescent labels which were not acquired for a given example. (B) The untrained deep network was (C) trained on the data A. (D) A z-stack of images of a novel scene. (E) The trained network, C, is used to predict fluorescence labels learned from A for each pixel in the novel images, D. |
In the course of this work we developed a new neural network composed of three kinds of basic building-blocks, inspired by the modular design of
Inception: an
in-scale configuration which does not change the spatial scaling of the features, a
down-scale configuration which doubles the spatial scaling, and an
up-scale configuration which halves it. This lets us break the hard problem of network architecture design into two easier problems: the arrangement of the building-blocks (macro-architecture), and the design of the building-blocks themselves (micro-architecture). We solved the first problem using design principles discussed in the paper, and the second via an automated search powered by
Google Hypertune.
To make sure our method was sound, we validated our model using data from an Alphabet lab as well as two external partners:
Steve Finkbeiner's lab at the Gladstone Institutes, and the
Rubin Lab at Harvard. These data spanned three transmitted light imaging modalities (
bright-field,
phase-contrast, and
differential interference contrast) and three culture types (human
motor neurons derived from
induced pluripotent stem cells, rat
cortical cultures, and human breast cancer cells). We found that our method can accurately predict several labels including those for nuclei, cell type (e.g. neural), and cell state (e.g. cell death). The following figure shows the model’s predictions alongside the transmitted light input and fluorescence ground-truth for our motor neuron example.
 |
| Animation showing the same cells in transmitted light and fluorescence imaging, along with predicted fluorescence labels from our model. Outset 2 shows the model predicts the correct labels despite the artifact in the input image. Outset 3 shows the model infers these processes are axons, possibly because of their distance from the nearest cells. Outset 4 shows the model sees the hard-to-see cell at the top, and correctly identifies the object at the left as DNA-free cell debris. |
Try it for yourself!We’ve
open sourced our model, along with our full dataset, code for training and inference, and an example. We’re pleased to report that new labels can be learned with minimal additional training data: in the paper and example code, we show a new label may be learned from a
single image. This is due to a phenomenon called
transfer learning, where a model can learn a new task more quickly and using less training data if it has already mastered similar tasks.
We hope the ability to generate labeled, useful, images without modifying cells will open up completely new kinds of experiments in biology and medicine. If you’re excited to try this technology in your own work, please read the paper or
check out the code!
AcknowledgementsWe thank the Google Accelerated Science team for originating and developing this project and its publication, and additionally Kevin P. Murphy for supporting its publication. We thank Mike Ando, Youness Bennani, Amy Chung-Yu Chou, Jason Freidenfelds, Jason Miller, Kevin P. Murphy, Philip Nelson, Patrick Riley, and Samuel Yang for ideas and editing help with this post. This study was supported by NINDS (NS091046, NS083390, NS101995), the NIH’s National Institute on Aging (AG065151, AG058476), the NIH’s National Human Genome Research Institute (HG008105), Google, the ALS Association, and the Michael J. Fox Foundation.