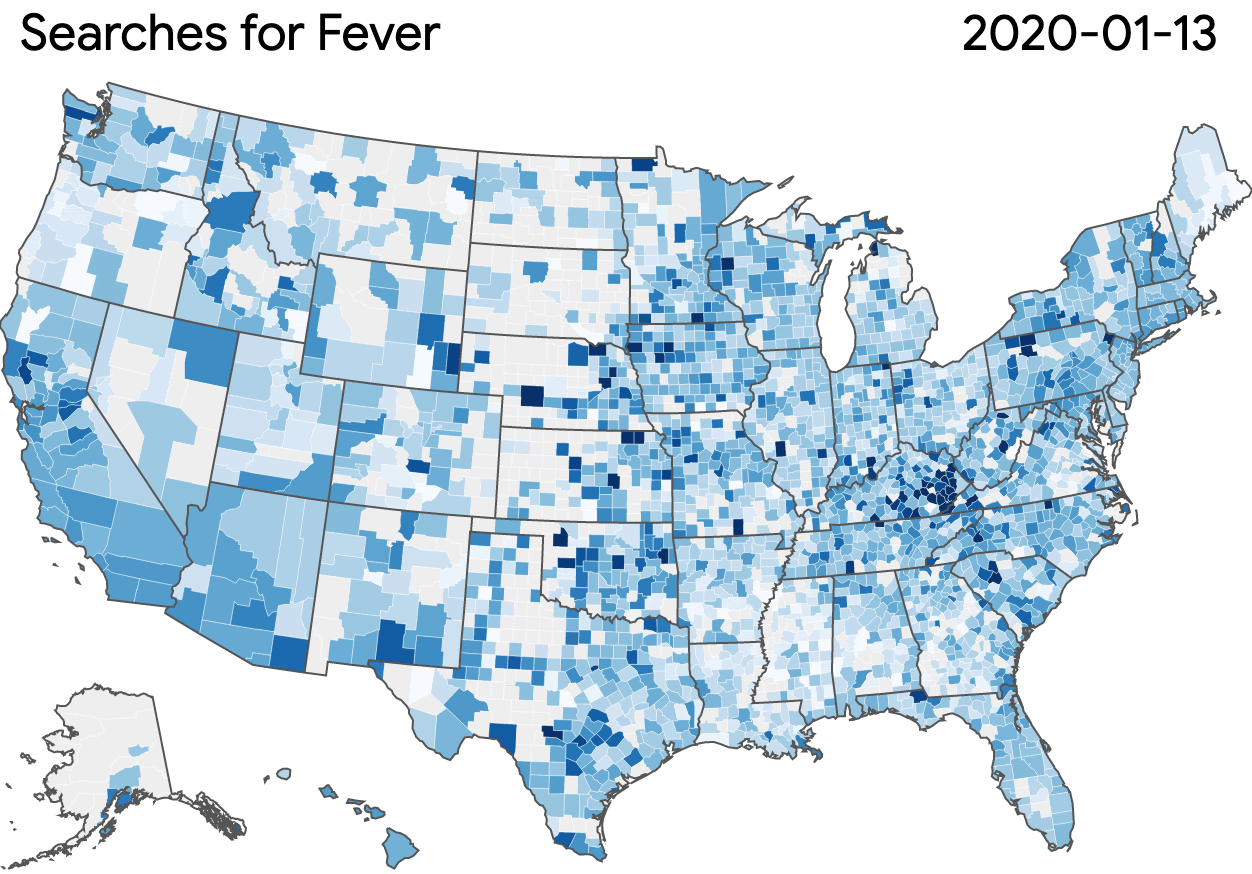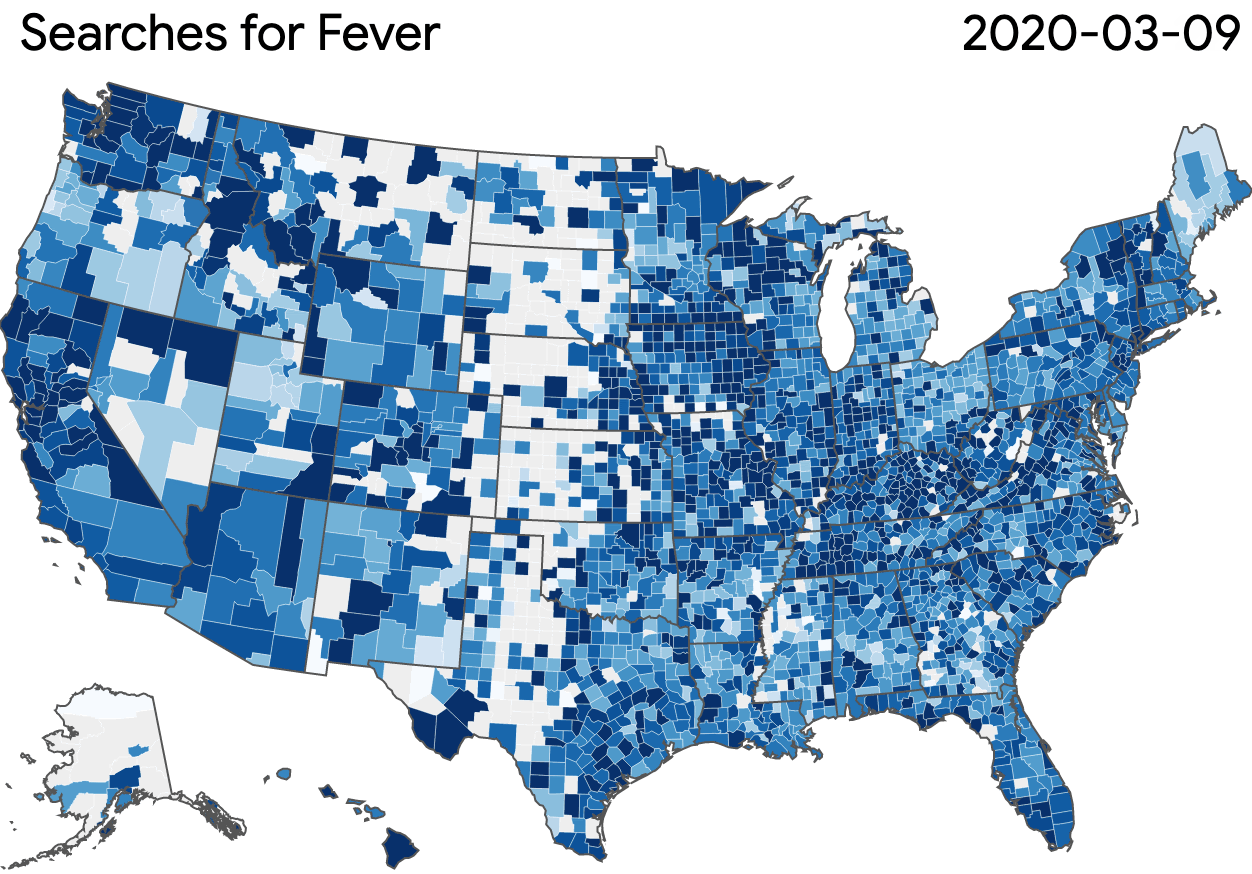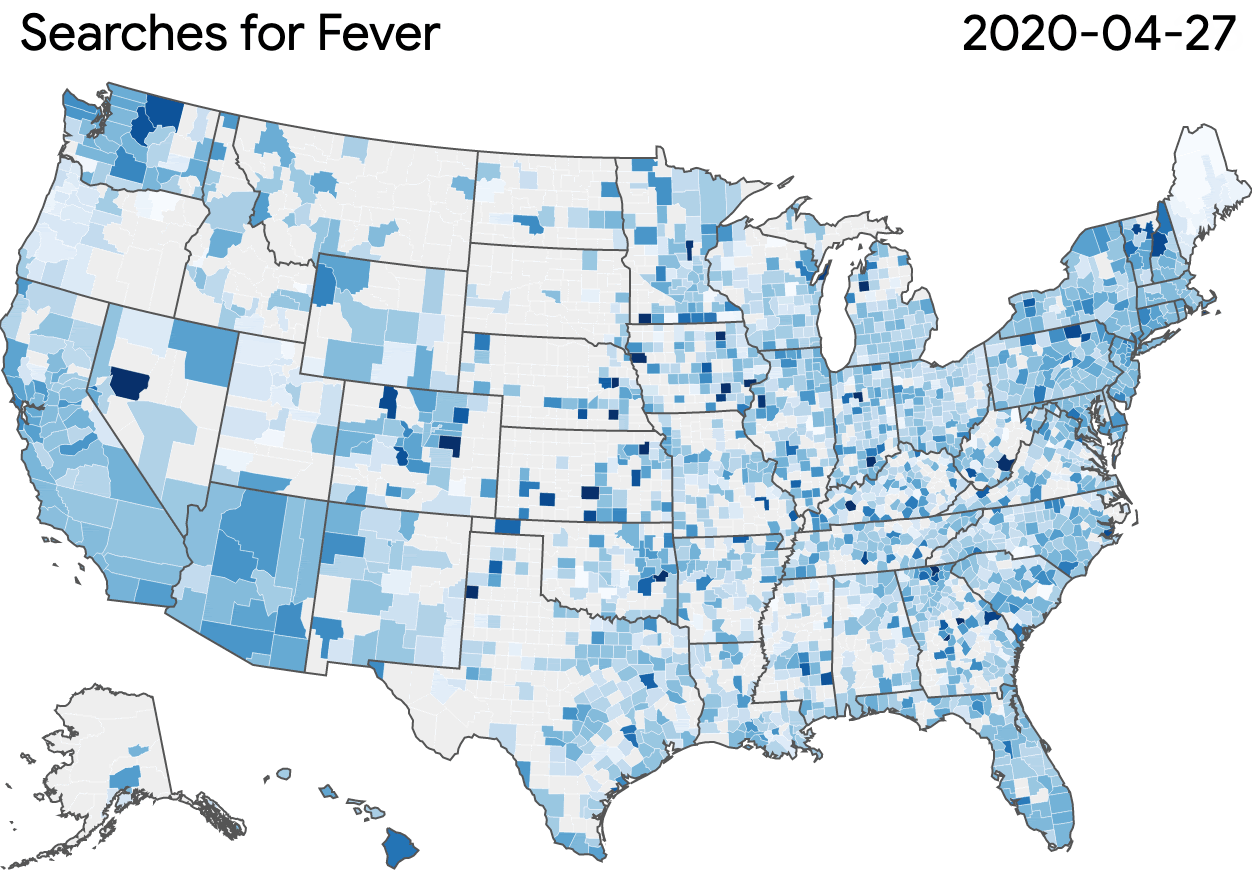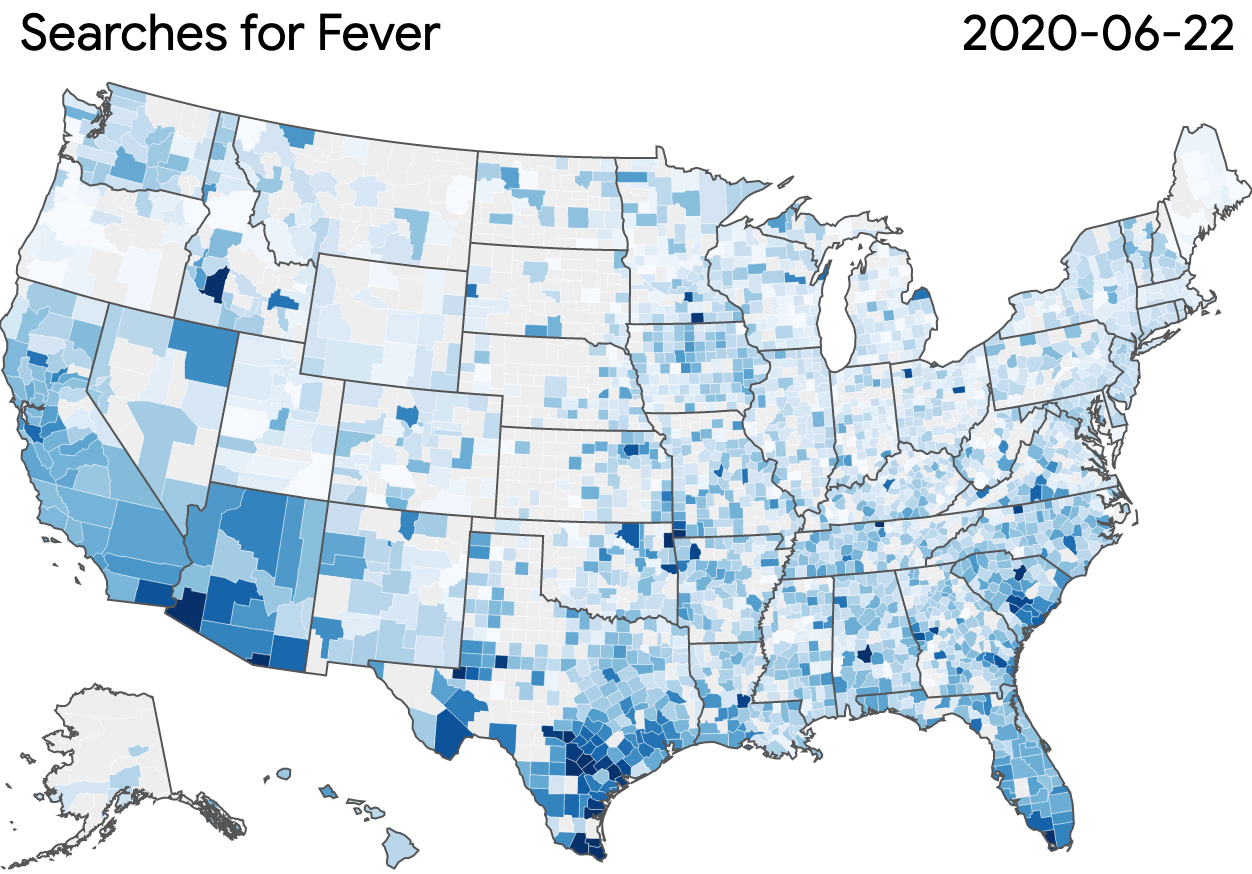
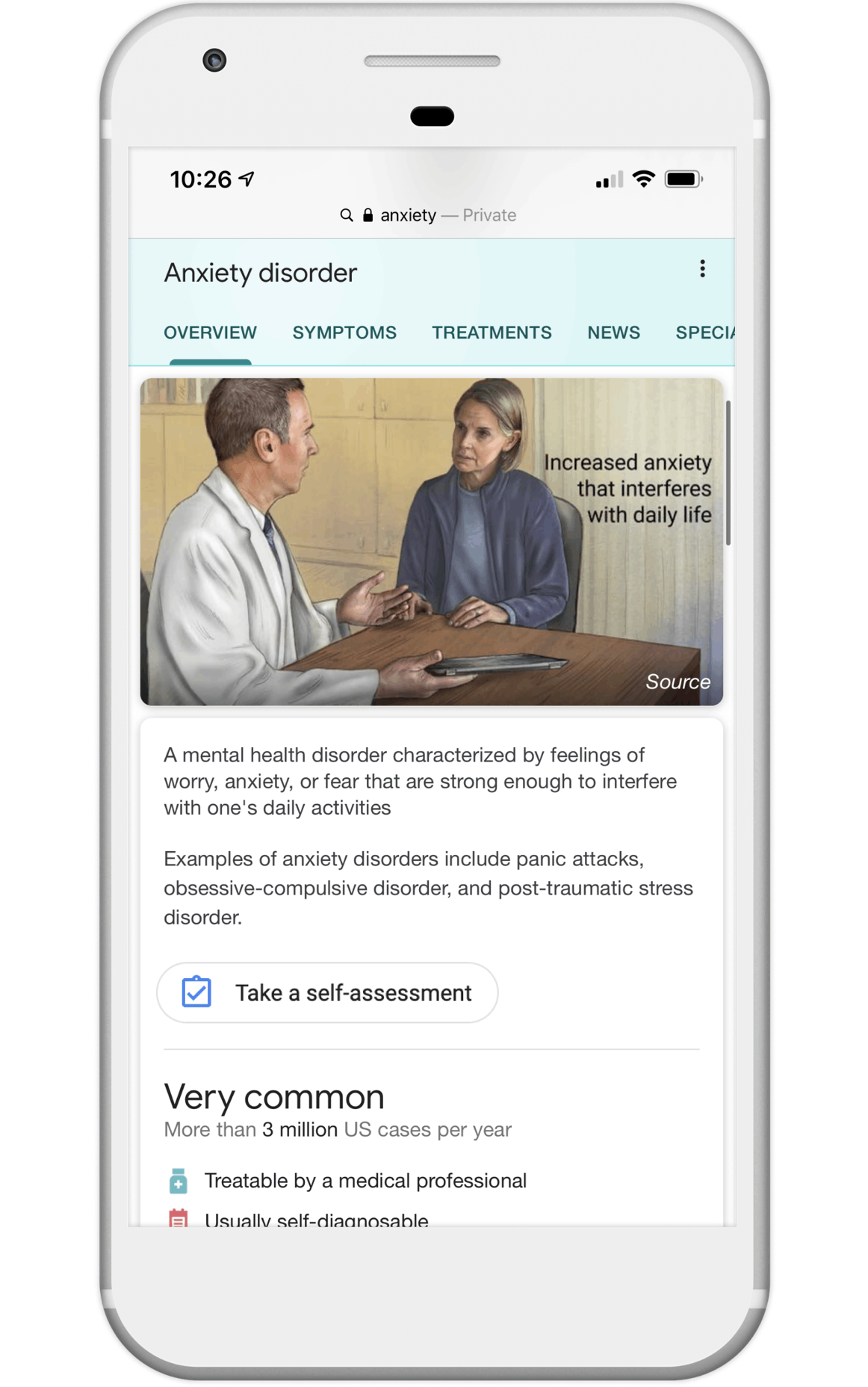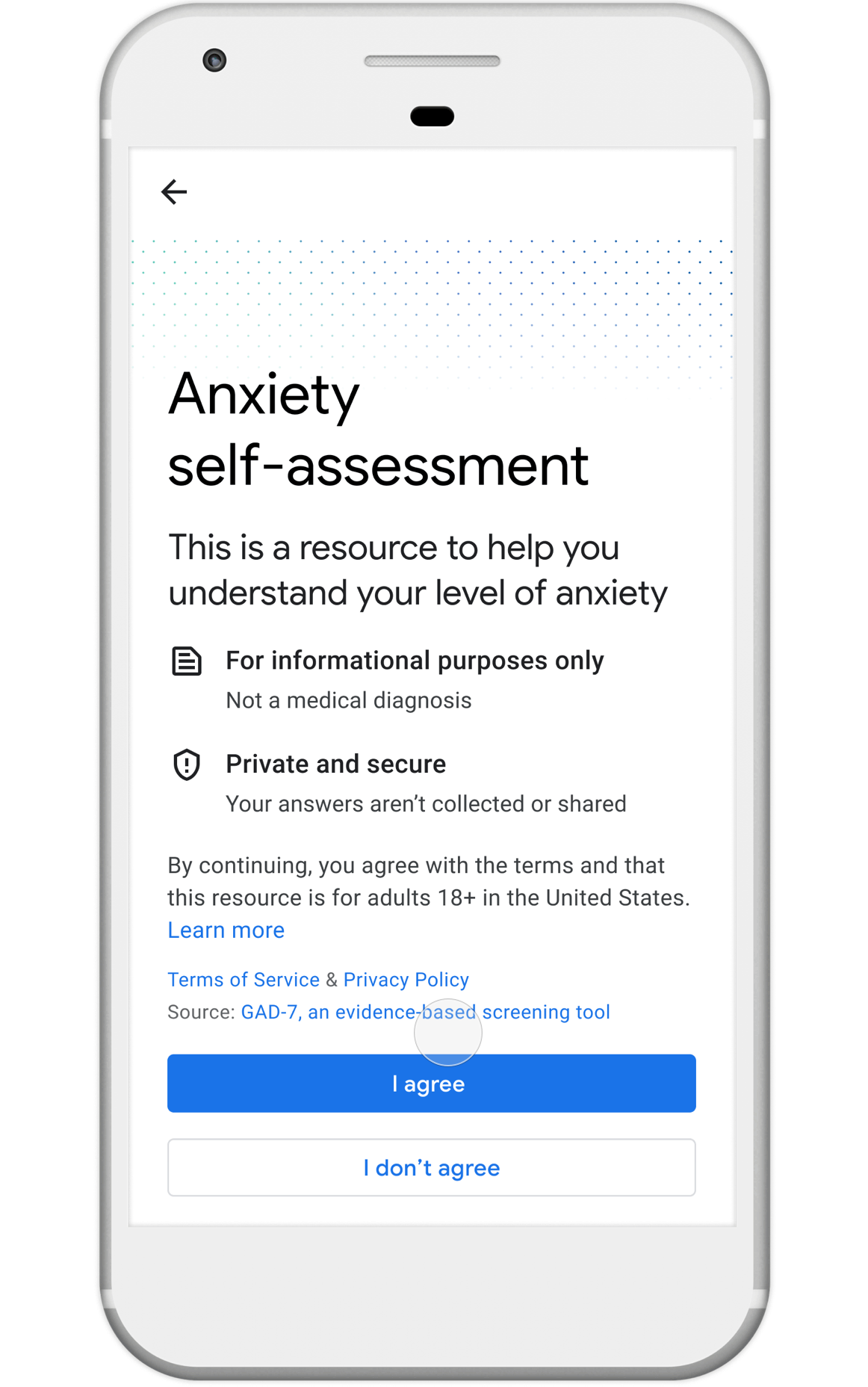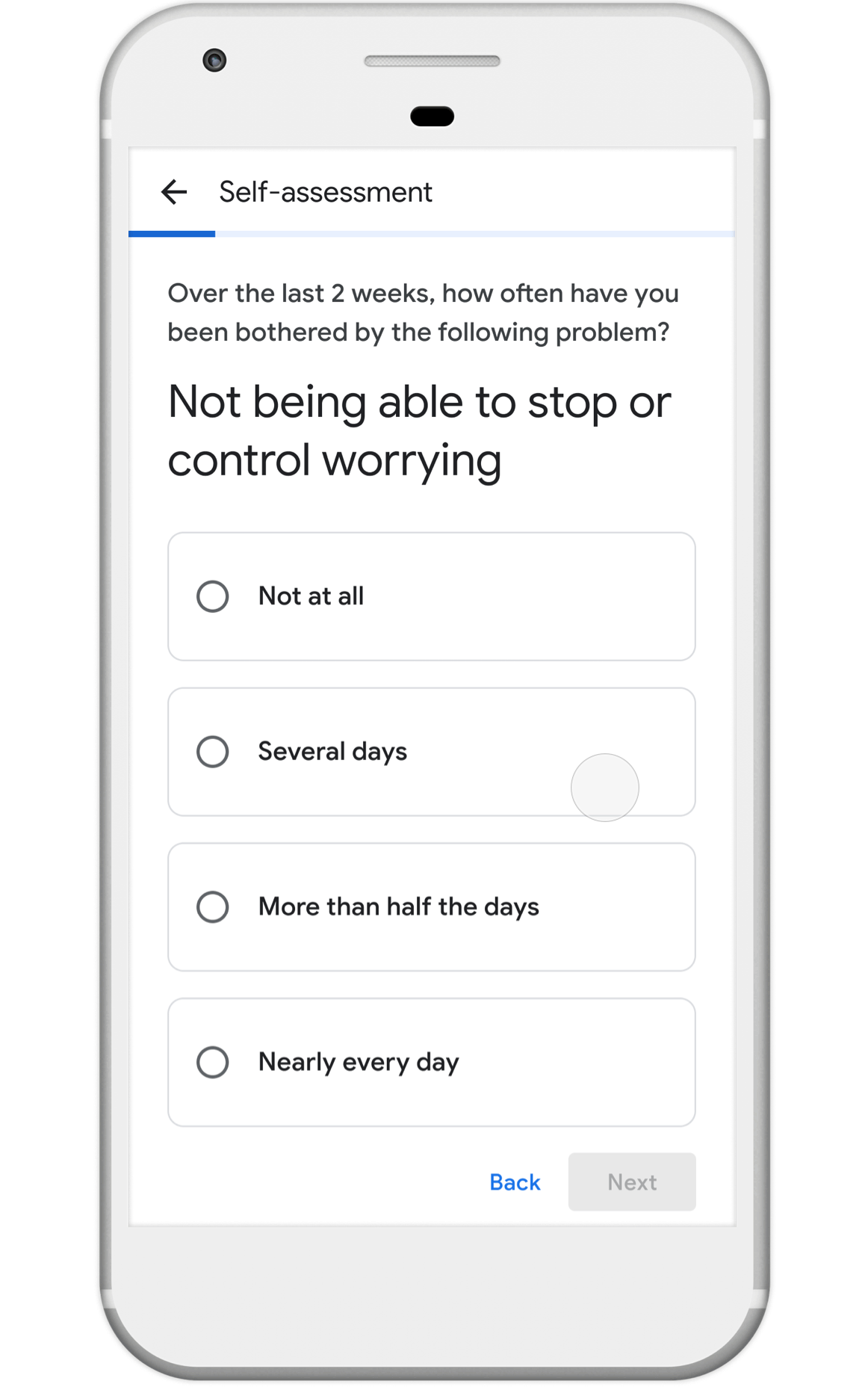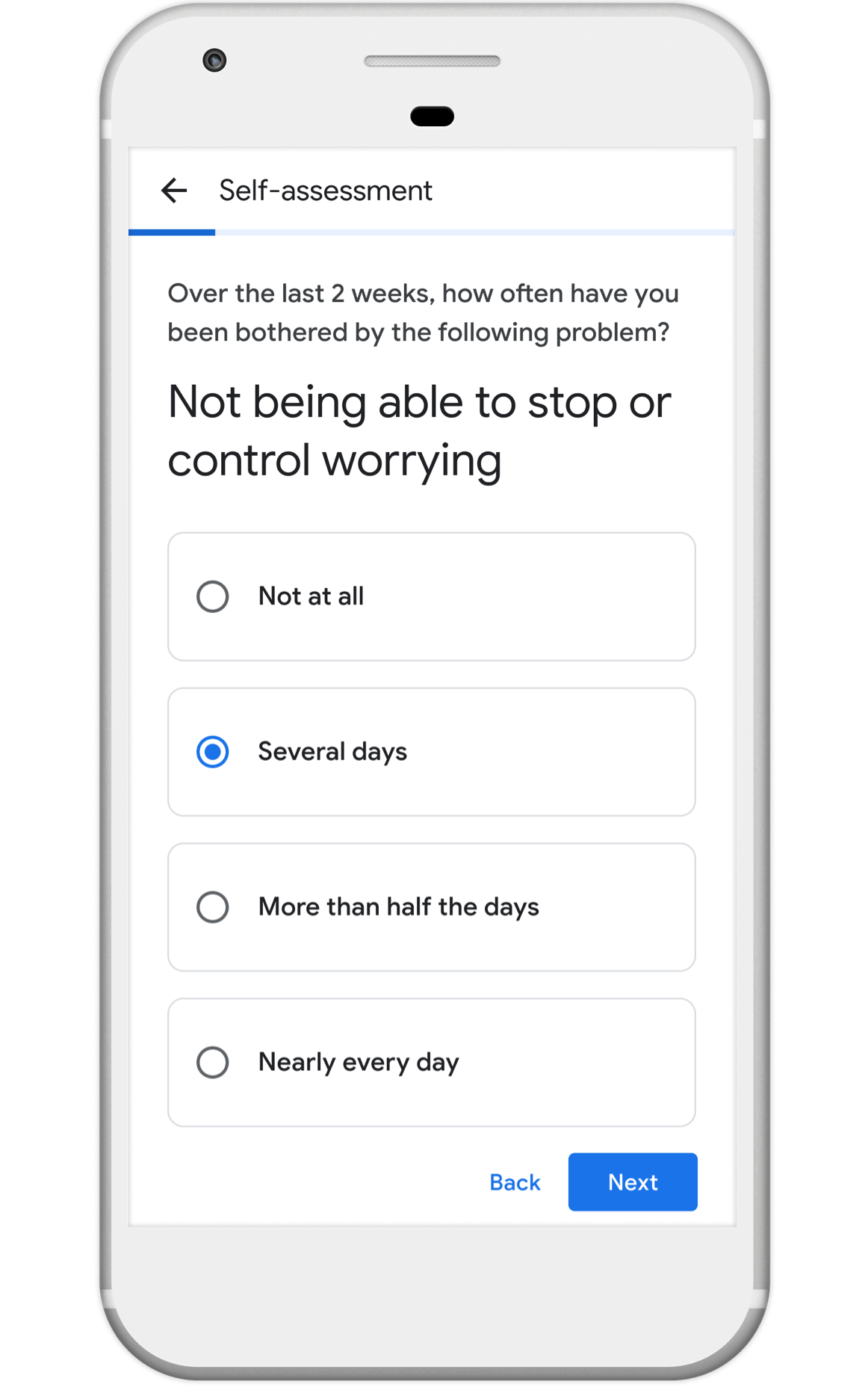
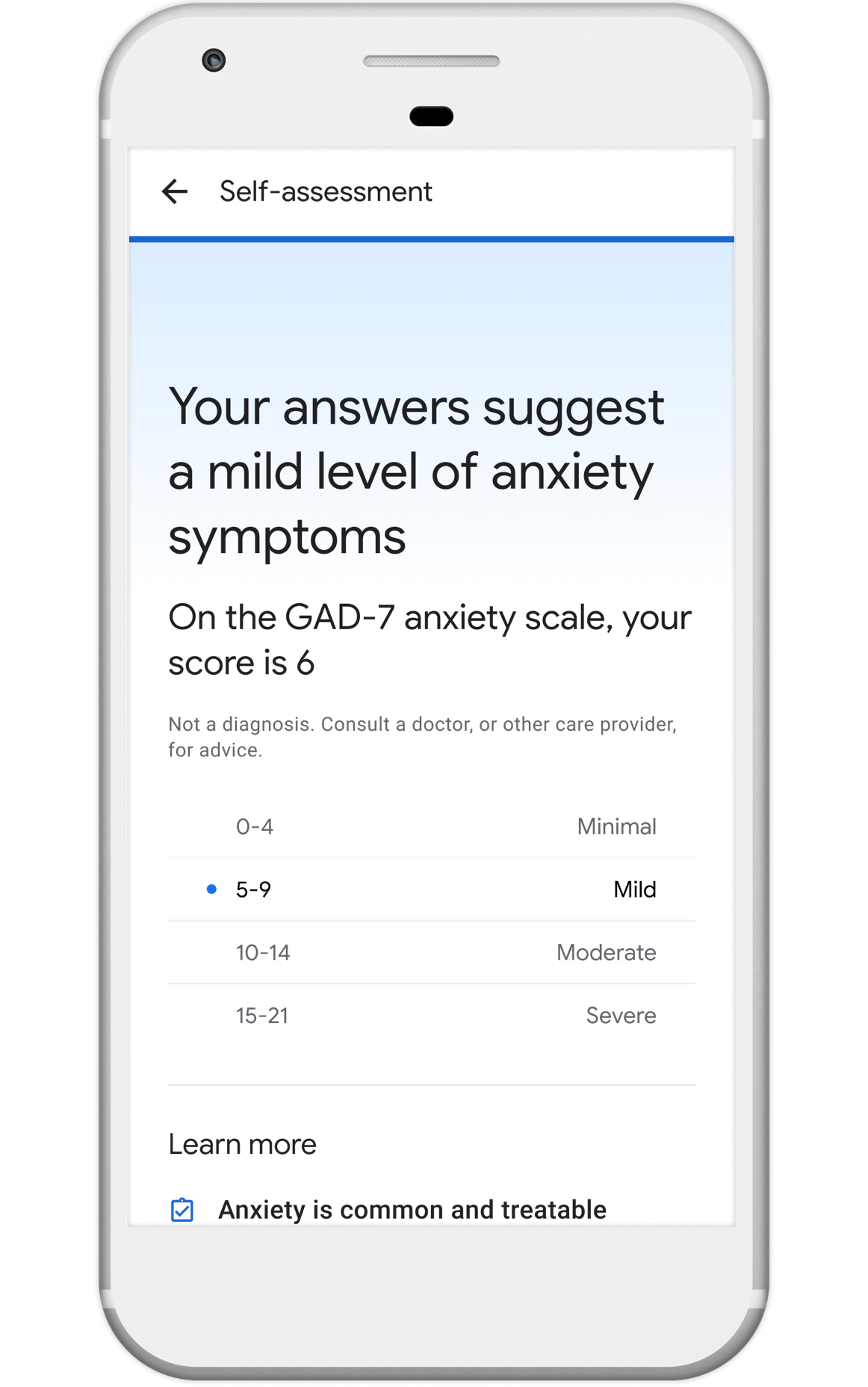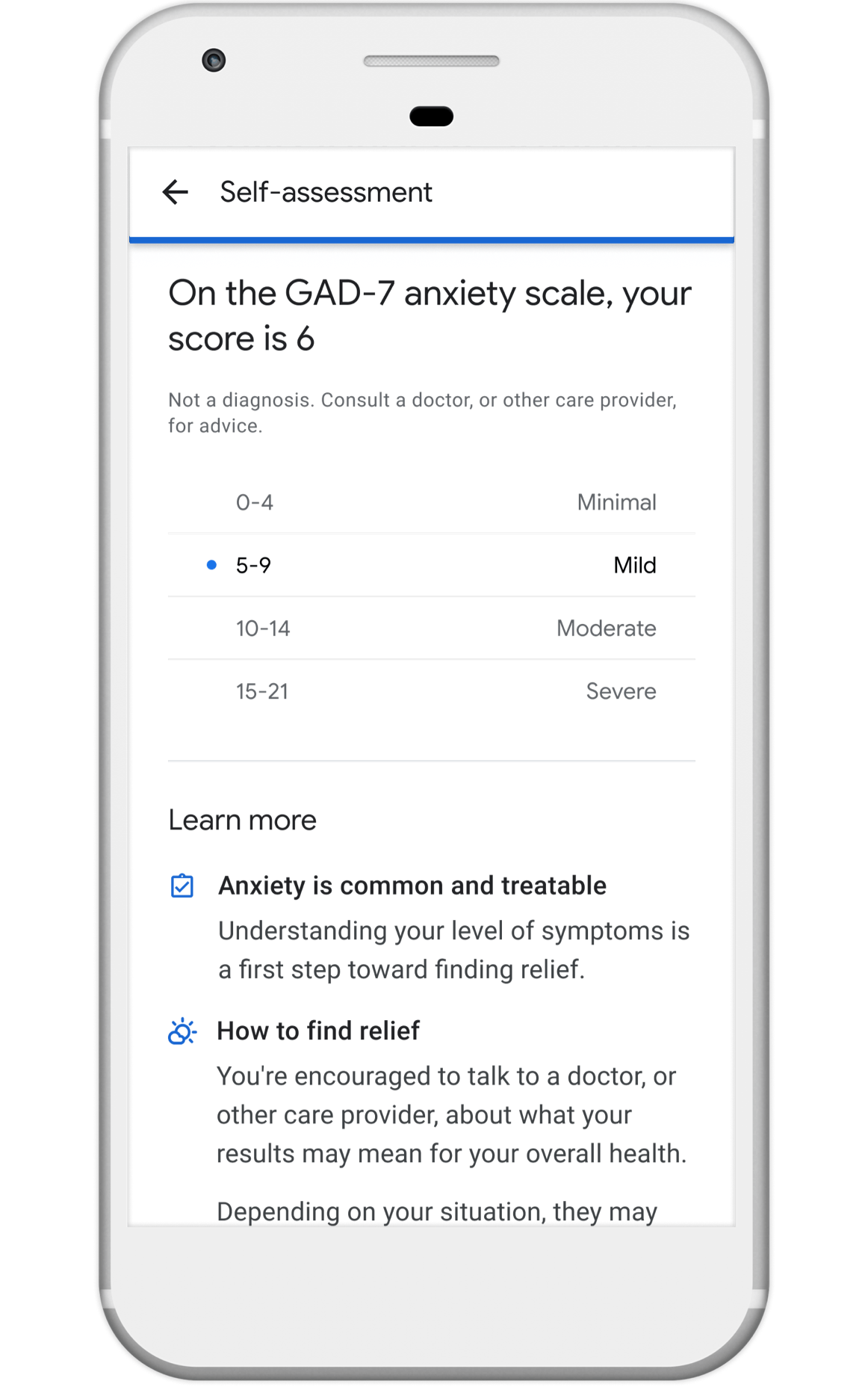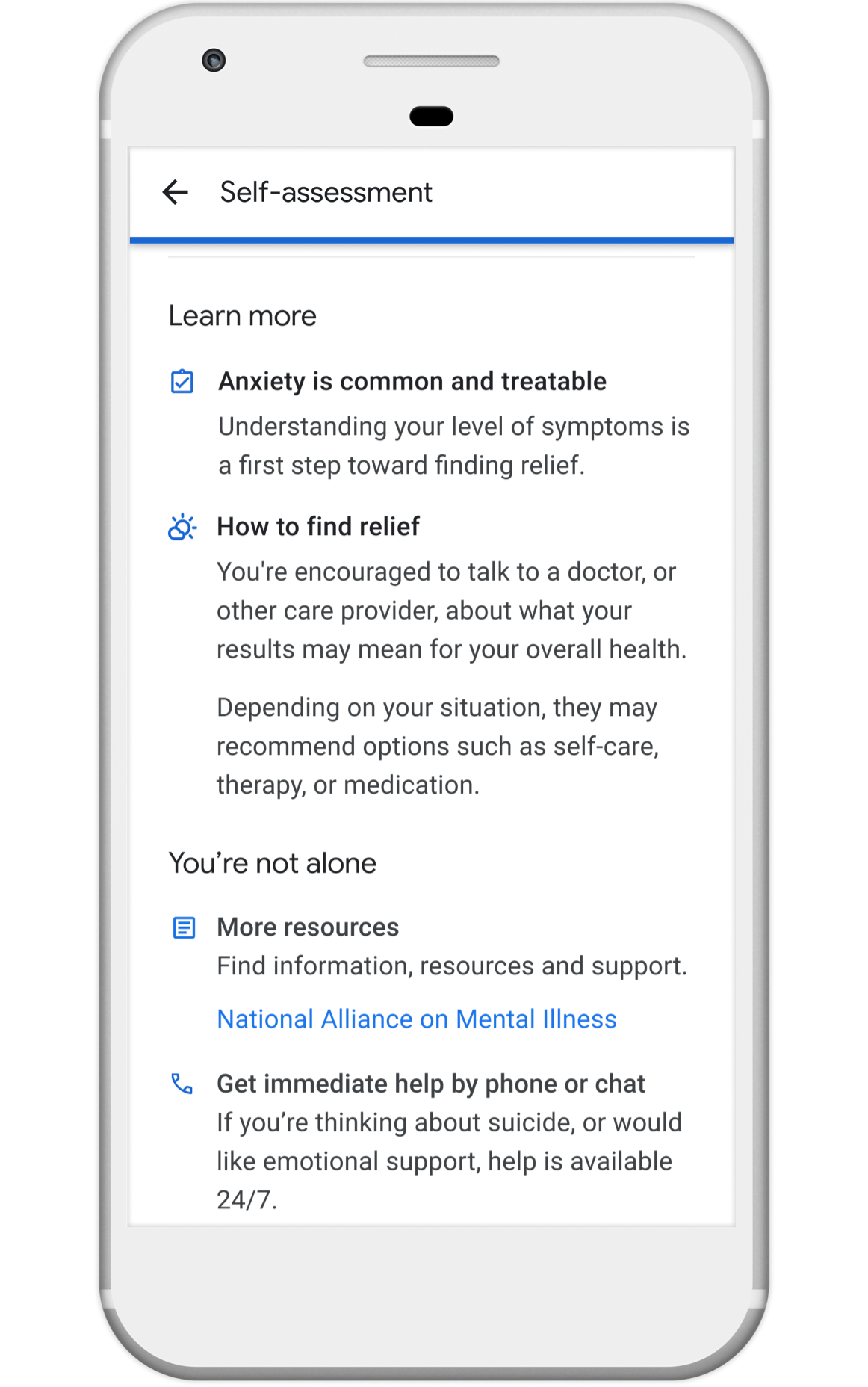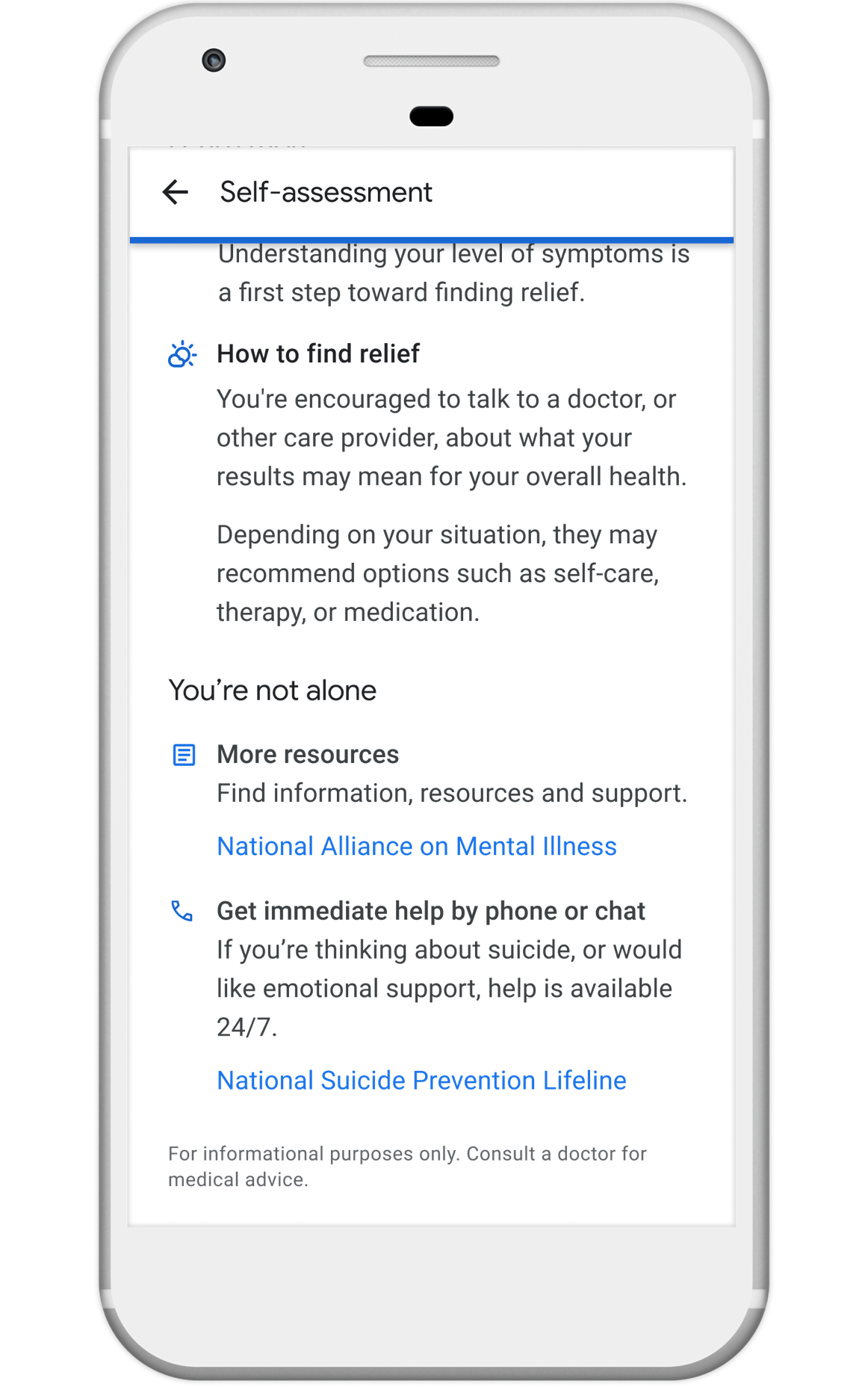
Nonprofits, universities and other academic institutions around the world are turning to artificial intelligence (AI) and data analytics to help us better understand COVID-19 and its impact on communities—especially vulnerable populations and healthcare workers. To support this work, Google.org is giving more than $8.5 million to 31 organizations around the world to aid in COVID-19 response. Three of these organizations will also receive the pro-bono support of Google.org Fellowship teams.
This funding is part of Google.org’s $100 million commitment to COVID-19 relief and focuses on four key areas where new information and action is needed to help mitigate the effects of the pandemic.
Monitoring and forecasting disease spread
Understanding the spread of COVID-19 is critical to informing public health decisions and lessening its impact on communities. We’re supporting the development of data platforms to help model disease and projects that explore the use of diverse public datasets to more accurately predict the spread of the virus.
Improving health equity and minimizing secondary effects of the pandemic
COVID-19 has had a disproportionate effect on vulnerable populations. To address health disparities and drive equitable outcomes, we’re supporting efforts to map the social and environmental drivers of COVID-19 impact, such as race, ethnicity, gender and socioeconomic status. In addition to learning more about the immediate health effects of COVID-19, we’re also supporting work that seeks to better understand and reduce the long-term, indirect effects of the virus—ranging from challenges with mental health to delays in preventive care.
Slowing transmission by advancing the science of contact tracing and environmental sensing
Contact tracing is a valuable tool to slow the spread of disease. Public health officials around the world are using digital tools to help with contact tracing. Google.org is supporting projects that advance science in this important area, including research investigating how to improve exposure risk assessments while preserving privacy and security. We’re also supporting related research to understand how COVID-19 might spread in public spaces, like transit systems.
Supporting healthcare workers
Whether it’s working to meet the increased demand for acute patient care, adapting to rapidly changing protocols or navigating personal mental and physical wellbeing, healthcare workers face complex challenges on the frontlines. We’re supporting organizations that are focused on helping healthcare workers quickly adopt new protocols, deliver more efficient care, and better serve vulnerable populations.
Together, these organizations are helping make the community’s response to the pandemic more advanced and inclusive, and we’re proud to support these efforts. You can find information about the organizations Google.org is supporting below.
Monitoring and forecasting disease spread
Carnegie Mellon University*: informing public health officials with interactive maps that display real-time COVID-19 data from sources such as web surveys and other publicly-available data.
Keio University: investigating the reliability of large-scale surveys in helping model the spread of COVID-19.
University College London:modeling the prevalence of COVID-19 and understanding its impact using publicly-available aggregated, anonymized search trends data.
Boston Children's Hospital, Oxford University, Northeastern University*: building a platform to support accurate and trusted public health data for researchers, public health officials and citizens.
Tel Aviv University: developing simulation models using synthetic data to investigate the spread of COVID-19 in Israel.
Kampala International University, Stanford University, Leiden University, GO FAIR: implementing data sharing standards and platforms for disease modeling for institutions across Uganda, Ethiopia, Nigeria, Kenya, Tunisia and Zimbabwe.
Improving health equity and minimizing secondary effects of the pandemic
Morehouse School of Medicine’s Satcher Health Leadership Institute*: developing an interactive, public-facing COVID-19 Health Equity Tracker of the United States.
Florida A&M University, Shaw University: examining structural social determinants of health and the disproportionate impact of COVID-19 in communities of color in Florida and North Carolina.
Boston University School of Public Health:investigating the drivers of racial, ethnic and socioeconomic disparities in the causes and consequences of COVID-19, with a focus on Massachusetts.
University of North Carolina, Vanderbilt University:investigating molecular mechanisms underlying susceptibility to SARS-CoV-2 and variability in COVID-19 outcomes in Hispanic/Latinx populations.
Beth Israel Deaconess Medical Center: quantifying the impact of COVID-19 on healthcare not directly associated with the virus, such as delayed routine or preventative care.
Georgia Institute of Technology:investigating opportunities for vulnerable populations to find information related to COVID-19.
Cornell Tech:developing digital tools and resources for advocates and survivors of intimate partner violence during COVID-19.
University of Michigan School of Information: evaluating health equity impacts of the rapid virtualization of primary healthcare.
Indian Institute of Technology Gandhinagar: modeling the impact of air pollution on COVID-related secondary health exacerbations.
Cornell University, EURECOM:developing scalable and explainable methods for verifying claims and identifying misinformation about COVID-19.
Slowing transmission by advancing the science of contact tracing and environmental sensing
Arizona State University:applying federated analytics (a state-of-the-art, privacy-preserving analytic technique) to contact tracing, including an on-campus pilot.
Stanford University:applying sparse secure aggregation to detect emerging hotspots.
University of Virginia, Princeton University, University of Maryland:designing and analyzing effective digital contact tracing methods.
University of Washington: investigating environmental SARS-CoV-2 detection and filtration methods in bus lines and other public spaces.
Indian Institute of Science, Bengaluru:mitigating the spread of COVID-19 in India’s transit systems with rapid testing and modified commuter patterns.
TU Berlin, University of Luxembourg:using quantum mechanics and machine learning to understand the binding of SARS-CoV-2 spike protein to human cells—a key process in COVID-19 infection.
Supporting healthcare workers
Medic Mobile, Dimagi: developing data analytics tools to support frontline health workers in countries such as India and Kenya.
Global Strategies:developing software to support healthcare workers adopting COVID-19 protocols in underserved, rural populations in the U.S., including Native American communities.
C Minds:creating an open-source, AI-based support system for clinical trials related to COVID-19.
Hospital Israelita Albert Einstein:supporting and integrating community health workers and volunteers to help deliver mental health services and monitor outcomes in one of Brazil's most vulnerable communities.
Fiocruz Bahia, Federal University of Bahia:establishing an AI platform for research and information-sharing related to COVID-19 in Brazil.
RAD-AID:creating and managing a data lake for institutions in low- and middle-income countries to pool anonymized data and access AI tools.
Yonsei University College of Medicine: scaling and distributing decision support systems for patients and doctors to better predict hospitalization and intensive care needs due to COVID-19.
University of California Berkeley and Gladstone Institutes: developing rapid at-home CRISPR-based COVID-19 diagnostic tests using cell phone technology.
Fondazione Istituto Italiano di Tecnologia:enabling open-source access to anonymized COVID-19 chest X-ray and clinical data, and researching image analysis for early diagnosis and prognosis.
*Recipient of a Google.org Fellowship