Natural language enables flexible descriptive queries about images. The interaction between text queries and images grounds linguistic meaning in the visual world, facilitating a better understanding of object relationships, human intentions towards objects, and interactions with the environment. The research community has studied object-level visual grounding through a range of tasks, including referring expression comprehension, text-based localization, and more broadly object detection, each of which require different skills in a model. For example, object detection seeks to find all objects from a predefined set of classes, which requires accurate localization and classification, while referring expression comprehension localizes an object from a referring text and often requires complex reasoning on prominent objects. At the intersection of the two is text-based localization, in which a simple category-based text query prompts the model to detect the objects of interest.
Due to their dissimilar task properties, referring expression comprehension, detection, and text-based localization are mostly studied through separate benchmarks with most models only dedicated to one task. As a result, existing models have not adequately synthesized information from the three tasks to achieve a more holistic visual and linguistic understanding. Referring expression comprehension models, for instance, are trained to predict one object per image, and often struggle to localize multiple objects, reject negative queries, or detect novel categories. In addition, detection models are unable to process text inputs, and text-based localization models often struggle to process complex queries that refer to one object instance, such as “Left half sandwich.” Lastly, none of the models can generalize sufficiently well beyond their training data and categories.
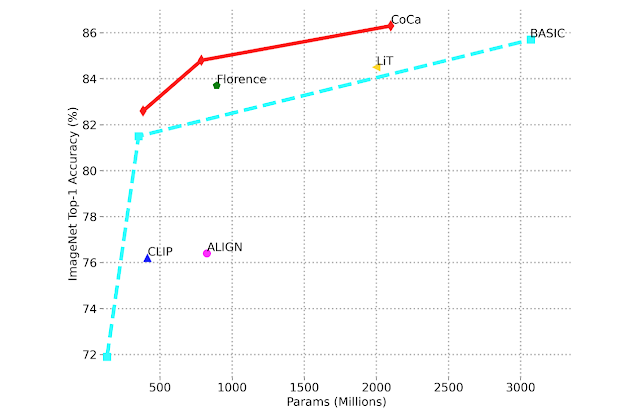
To address these limitations, we are presenting “FindIt: Generalized Localization with Natural Language Queries” at ECCV 2022. Here we propose a unified, general-purpose and multitask visual grounding model, called FindIt, that can flexibly answer different types of grounding and detection queries. Key to this architecture is a multi-level cross-modality fusion module that can perform complex reasoning for referring expression comprehension and simultaneously recognize small and challenging objects for text-based localization and detection. In addition, we discover that a standard object detector and detection losses are sufficient and surprisingly effective for all three tasks without the need for task-specific design and losses common in existing works. FindIt is simple, efficient, and outperforms alternative state-of-the-art models on the referring expression comprehension and text-based localization benchmarks, while being competitive on the detection benchmark.
 |
| FindIt is a unified model for referring expression comprehension (col. 1), text-based localization (col. 2), and the object detection task (col. 3). FindIt can respond accurately when tested on object types/classes not known during training, e.g. “Find the desk” (col. 4). Compared to existing baselines (MattNet and GPV), FindIt can perform these tasks well and in a single model. |
Multi-level Image-Text Fusion
Different localization tasks are created with different semantic understanding objectives. For example, because the referring expression task primarily references prominent objects in the image rather than small, occluded or faraway objects, low resolution images generally suffice. In contrast, the detection task aims to detect objects with various sizes and occlusion levels in higher resolution images. Apart from these benchmarks, the general visual grounding problem is inherently multiscale, as natural queries can refer to objects of any size. This motivates the need for a multi-level image-text fusion model for efficient processing of higher resolution images over different localization tasks.
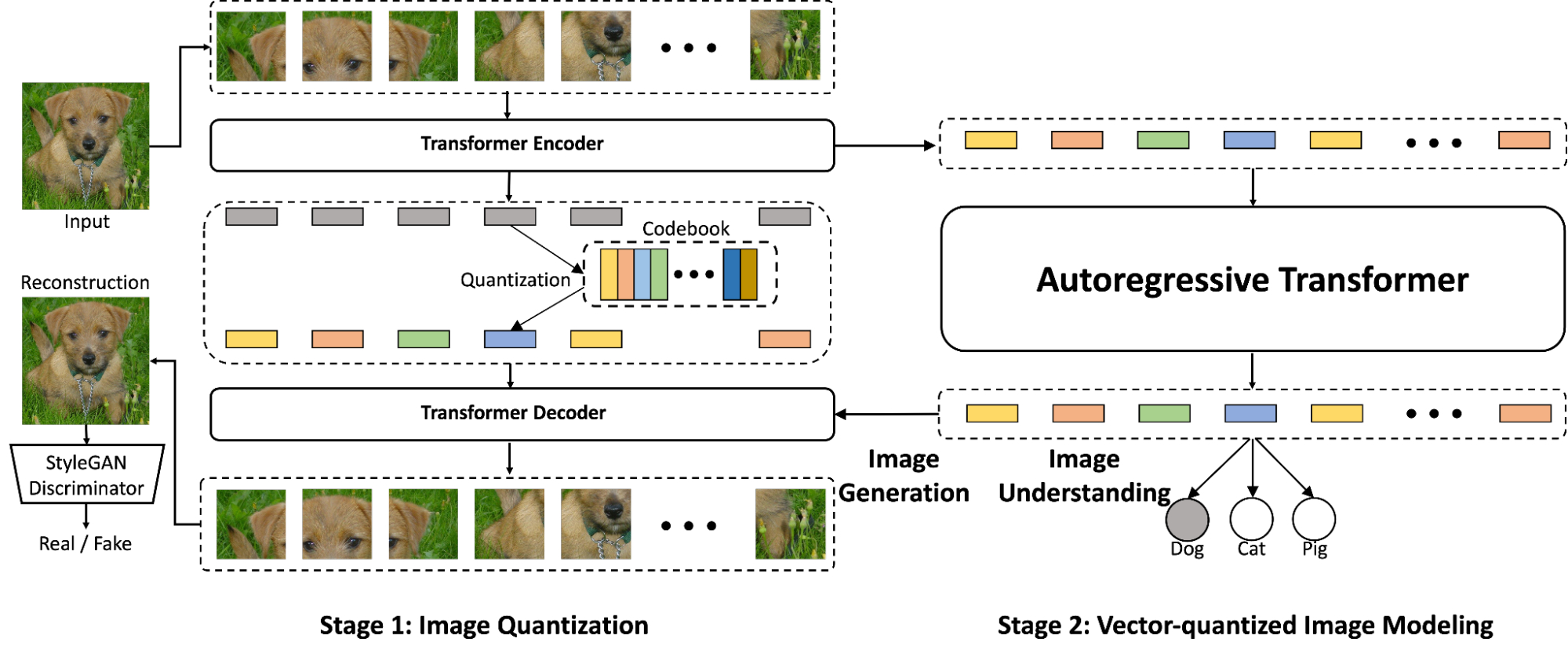
The premise of FindIt is to fuse the higher level semantic features using more expressive transformer layers, which can capture all-pair interactions between image and text. For the lower-level and higher-resolution features, we use a cheaper dot-product fusion to save computation and memory cost. We attach a detector head (e.g., Faster R-CNN) on top of the fused feature maps to predict the boxes and their classes.
 |
| FindIt accepts an image and a query text as inputs, and processes them separately in image/text backbones before applying the multi-level fusion. We feed the fused features to Faster R-CNN to predict the boxes referred to by the text. The feature fusion uses more expressive transformers at higher levels and cheaper dot-product at the lower levels. |
Multitask Learning
Apart from the multi-level fusion described above, we adapt the text-based localization and detection tasks to take the same inputs as the referring expression comprehension task. For the text-based localization task, we generate a set of queries over the categories present in the image. For any present category, the text query takes the form “Find the [object],” where [object] is the category name. The objects corresponding to that category are labeled as foreground and the other objects as background. Instead of using the aforementioned prompt, we use a static prompt for the detection task, such as “Find all the objects.”. We found that the specific choice of prompts is not important for text-based localization and detection tasks.
After adaptation, all tasks in consideration share the same inputs and outputs — an image input, a text query, and a set of output bounding boxes and classes. We then combine the datasets and train on the mixture. Finally, we use the standard object detection losses for all tasks, which we found to be surprisingly simple and effective.
Evaluation
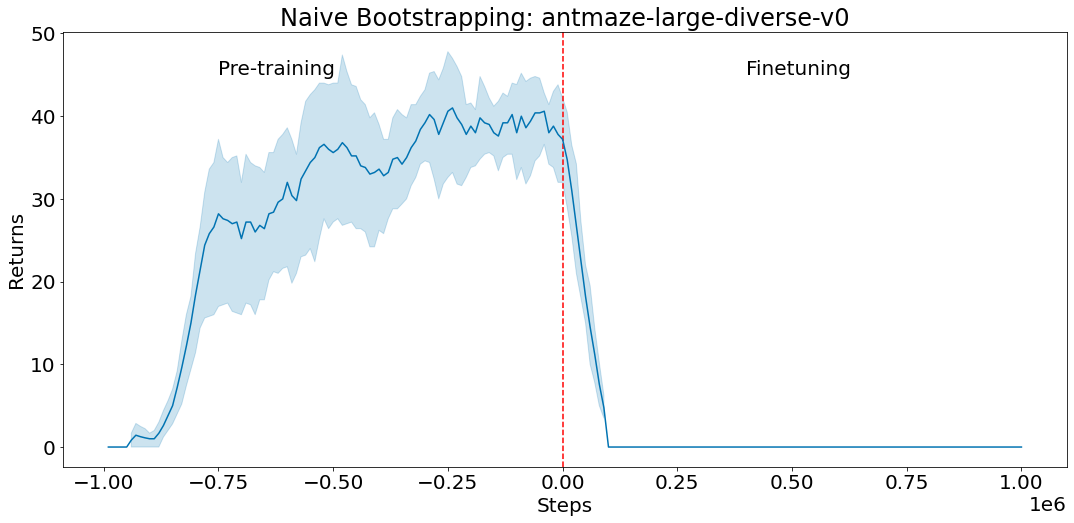
We apply FindIt to the popular RefCOCO benchmark for referring expression comprehension tasks. When only the COCO and RefCOCO dataset is available, FindIt outperforms the state-of-the-art-model on all tasks. In the settings where external datasets are allowed, FindIt sets a new state of the art by using COCO and all RefCOCO splits together (no other datasets). On the challenging Google and UMD splits, FindIt outperforms the state of the art by a 10% margin, which, taken together, demonstrate the benefits of multitask learning.
 |
| Comparison with the state of the art on the popular referring expression benchmark. FindIt is superior on both the COCO and unconstrained settings (additional training data allowed). |
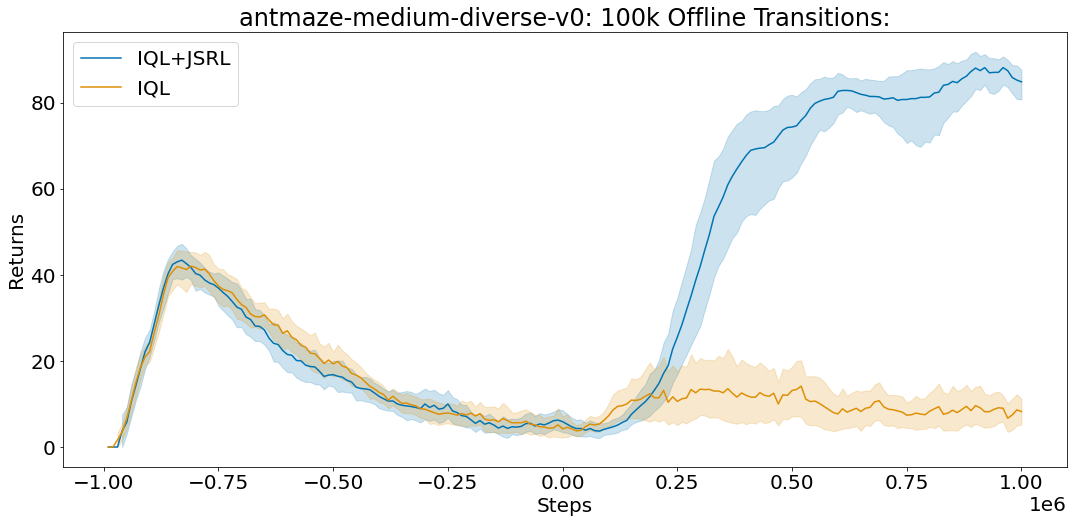
On the text-based localization benchmark, FindIt achieves 79.7%, higher than the GPV (73.0%), and Faster R-CNN baselines (75.2%). Please refer to the paper for more quantitative evaluation.
We further observe that FindIt generalizes better to novel categories and super-categories in the text-based localization task compared to competitive single-task baselines on the popular COCO and Objects365 datasets, shown in the figure below.
Efficiency
We also benchmark the inference times on the referring expression comprehension task (see Table below). FindIt is efficient and comparable with existing one-stage approaches while achieving higher accuracy. For fair comparison, all running times are measured on one GTX 1080Ti GPU.
| Model | Image Size | Backbone | Runtime (ms) | |||
| MattNet | 1000 | R101 | 378 | |||
| FAOA | 256 | DarkNet53 | 39 | |||
| MCN | 416 | DarkNet53 | 56 | |||
| TransVG | 640 | R50 | 62 | |||
| FindIt (Ours) | 640 | R50 | 107 | |||
| FindIt (Ours) | 384 | R50 | 57 |
Conclusion
We present Findit, which unifies referring expression comprehension, text-based localization, and object detection tasks. We propose multi-scale cross-attention to unify the diverse localization requirements of these tasks. Without any task-specific design, FindIt surpasses the state of the art on referring expression and text-based localization, shows competitive performance on detection, and generalizes better to out-of-distribution data and novel classes. All of these are accomplished in a single, unified, and efficient model.
Acknowledgements
This work is conducted by Weicheng Kuo, Fred Bertsch, Wei Li, AJ Piergiovanni, Mohammad Saffar, and Anelia Angelova. We would like to thank Ashish Vaswani, Prajit Ramachandran, Niki Parmar, David Luan, Tsung-Yi Lin, and other colleagues at Google Research for their advice and helpful discussions. We would like to thank Tom Small for preparing the animation.


















.gif)