Posted by Alex D’Amour and Katherine Heller, Research Scientists, Google Research Machine learning (ML) models are being used more widely today than ever before and are becoming increasingly impactful. However, they often exhibit unexpected behavior when they are used in real-world domains. For example, computer vision models can exhibit surprising sensitivity to irrelevant features, while natural language processing models can depend unpredictably on demographic correlations not directly indicated by the text. Some reasons for these failures are well-known: for example, training ML models on poorly curated data, or training models to solve prediction problems that are structurally mismatched with the application domain. Yet, even when these known problems are handled, model behavior can still be inconsistent in deployment, varying even between training runs.
In “Underspecification Presents Challenges for Credibility in Modern Machine Learning”, to be published in the Journal of Machine Learning Research, we show that a key failure mode especially prevalent in modern ML systems is underspecification. The idea behind underspecification is that while ML models are validated on held-out data, this validation is often insufficient to guarantee that the models will have well-defined behavior when they are used in a new setting. We show that underspecification appears in a wide variety of practical ML systems and suggest some strategies for mitigation.
Underspecification
ML systems have been successful largely because they incorporate validation of the model on held-out data to ensure high performance. However, for a fixed dataset and model architecture, there are often many distinct ways that a trained model can achieve high validation performance. But under standard practice, models that encode distinct solutions are often treated as equivalent because their held-out predictive performance is approximately equivalent.
Importantly, the distinctions between these models do become clear when they are measured on criteria beyond standard predictive performance, such as fairness or robustness to irrelevant input perturbations. For example, among models that perform equally well on standard validations, some may exhibit greater performance disparities between social groups than others, or rely more heavily on irrelevant information. These differences, in turn, can translate to real differences in behavior when the model is used in real-world scenarios.
Underspecification refers to this gap between the requirements that practitioners often have in mind when they build an ML model, and the requirements that are actually enforced by the ML pipeline (i.e., the design and implementation of a model). An important consequence of underspecification is that even if the pipeline could in principle return a model that meets all of these requirements, there is no guarantee that in practice the model will satisfy any requirement beyond accurate prediction on held-out data. In fact, the model that is returned may have properties that instead depend on arbitrary or opaque choices made in the implementation of the ML pipeline, such as those arising from random initialization seeds, data ordering, hardware, etc. Thus, ML pipelines that do not include explicit defects may still return models that behave unexpectedly in real-world settings.
Identifying Underspecification in Real Applications
In this work, we investigated concrete implications of underspecification in the kinds of ML models that are used in real-world applications. Our empirical strategy was to construct sets of models using nearly identical ML pipelines, to which we only applied small changes that had no practical effect on standard validation performance. Here, we focused on the random seed used to initialize training and determine data ordering. If important properties of the model can be substantially influenced by these changes, it indicates that the pipeline does not fully specify this real-world behavior. In every domain where we conducted this experiment, we found that these small changes induced substantial variation on axes that matter in real-world use.
Underspecification in Computer Vision
As an example, consider underspecification and its relationship to robustness in computer vision. A central challenge in computer vision is that deep models often suffer from brittleness under distribution shifts that humans do not find challenging. For instance, image classification models that perform well on the ImageNet benchmark are known to perform poorly on benchmarks like ImageNet-C, which apply common image corruptions, such as pixelization or motion blur, to the standard ImageNet test set.
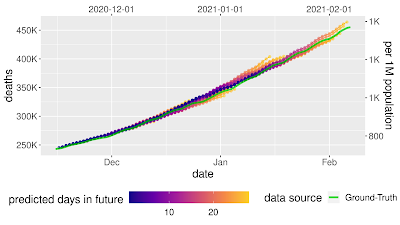
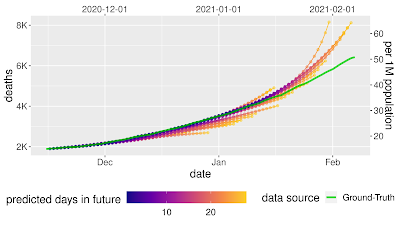
In our experiment, we showed that model sensitivity to these corruptions is underspecified by standard pipelines. Following the strategy discussed above, we generated fifty ResNet-50 image classification models using the same pipeline and the same data. The only difference between these models was the random seed used in training. When evaluated on the standard ImageNet validation set, these models achieved practically equivalent performance. However, when the models were evaluated on different test sets in the ImageNet-C benchmark (i.e., on corrupted data), performance on some tests varied by orders of magnitude more than on standard validations. This pattern persisted for larger-scale models that were pre-trained on much larger datasets (e.g., a BiT-L model pre-trained on the 300 million image JFT-300M dataset). For these models, varying the random seed at the fine-tuning stage of training produced a similar pattern of variations.
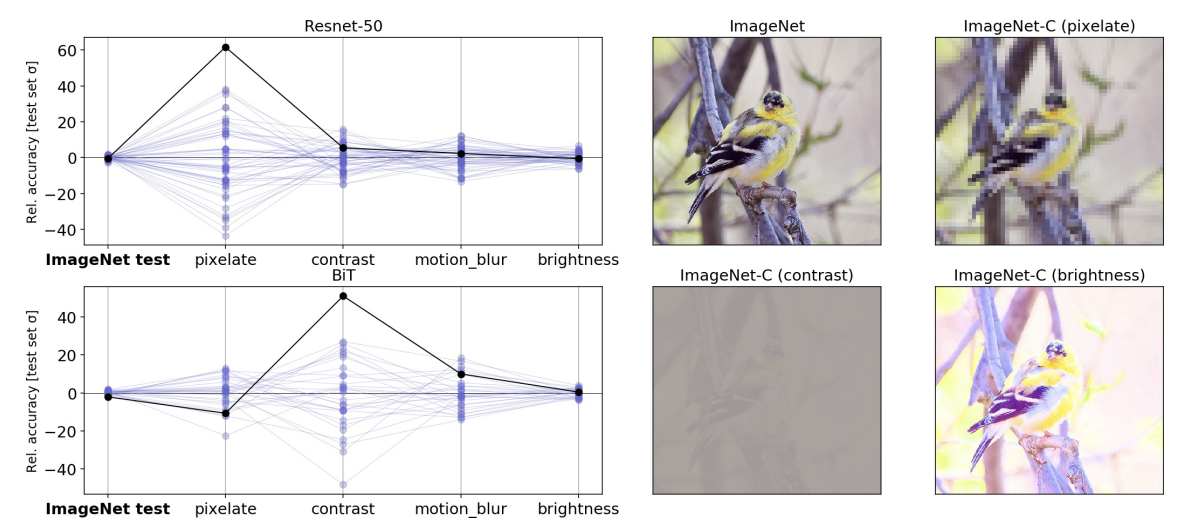 |
| Left: Parallel axis plots showing the variation in accuracy between identical, randomly initialized ResNet-50 models on strongly corrupted ImageNet-C data. Lines represent the performance of each model in the ensemble on classification tasks using uncorrupted test data, as well as corrupted data (pixelation, contrast, motion blur, and brightness). Given values are the deviation in accuracy from the ensemble mean, scaled by the standard deviation of accuracies on the “clean” ImageNet test set. The solid black line highlights the performance of an arbitrarily selected model to show how performance on one test may not be a good indication of performance on others. Right: Example images from the standard ImageNet test set, with corrupted versions from the ImageNet-C benchmark. |
We also showed that underspecification can have practical implications in special-purpose computer vision models built for medical imaging, where deep learning models have shown great promise. We considered two research pipelines intended as precursors for medical applications: one ophthalmology pipeline for building models that detect diabetic retinopathy and referable diabetic macular edema from retinal fundus images, and one dermatology pipeline for building models to recognize common dermatological conditions from photographs of skin. In our experiments, we considered pipelines that were validated only on randomly held-out data.
We then stress-tested models produced by these pipelines on practically important dimensions. For the ophthalmology pipeline, we tested how models trained with different random seeds performed when applied to images taken from a new camera type not encountered during training. For the dermatology pipeline, the stress test was similar, but for patients with different estimated skin types (i.e., non-dermatologist evaluation of tone and response to sunlight). In both cases, we found that standard validations were not enough to fully specify the trained model’s performance on these axes. In the ophthalmology application, the random seed used in training induced wider variability in performance on a new camera type than would have been expected from standard validations, and in the dermatology application, the random seed induced similar variation in performance in skin-type subgroups, even though the overall performance of the models was stable across seeds.
These results reiterate that standard hold-out testing alone is not sufficient to ensure acceptable model behavior in medical applications, underscoring the need for expanded testing protocols for ML systems intended for application in the medical domain. In the medical literature, such validations are termed "external validation" and have historically been part of reporting guidelines such as STARD and TRIPOD. These are being emphasized in updates such as STARD-AI and TRIPOD-AI. Finally, as part of regulated medical device development processes (see, e.g., US and EU regulations), there are other forms of safety and performance related considerations, such as mandatory compliance to standards for risk management, human factors engineering, clinical validations and accredited body reviews, that aim to ensure acceptable medical application performance.
 |
| Relative variability of medical imaging models on stress tests, using the same conventions as the figure above. Top left: Variation in AUC between diabetic retinopathy classification models trained using different random seeds when evaluated on images from different camera types. In this experiment, camera type 5 was not encountered during training. Bottom left: Variation in accuracy between skin condition classification models trained using different random seeds when evaluated on different estimated skin types (approximated by dermatologist-trained laypersons from retrospective photographs and potentially subject to labeling errors). Right: example images from the original test set (left) and the stress test set (right). |
Underspecification in Other Applications
The cases discussed above are a small subset of models that we probed for underspecification. Other cases we examined include:
- Natural Language Processing: We showed that on a variety of NLP tasks, underspecification affected how models derived from BERT-processed sentences. For example, depending on the random seed, a pipeline could produce a model that depends more or less on correlations involving gender (e.g., between gender and occupation) when making predictions.
- Acute Kidney Injury (AKI) prediction: We showed that underspecification affects reliance on operational versus physiological signals in AKI prediction models based on electronic health records.
- Polygenic Risk Scores (PRS): We showed that underspecification influences the ability for (PRS) models, which predict clinical outcomes based on patient genomic data, to generalize across different patient populations.
In each case, we showed that these important properties are left ill-defined by standard training pipelines, making them sensitive to seemingly innocuous choices.
Conclusion
Addressing underspecification is a challenging problem. It requires full specification and testing of requirements for a model beyond standard predictive performance. Doing this well needs full engagement with the context in which the model will be used, an understanding of how the training data were collected, and often, incorporation of domain expertise when the available data fall short. These aspects of ML system design are often underemphasized in ML research today. A key goal of this work is to show how underinvestment in this area can manifest concretely, and to encourage the development of processes for fuller specification and testing of ML pipelines.
Some important first steps in this area are to specify stress testing protocols for any applied ML pipeline that is meant to see real-world use. Once these criteria are codified in measurable metrics, a number of different algorithmic strategies may be useful for improving them, including data augmentation, pretraining, and incorporation of causal structure. It should be noted, however, that ideal stress testing and improvement processes will usually require iteration: both the requirements for ML systems, and the world in which they are used, are constantly changing.
Acknowledgements
We would like to thank all of our co-authors, Dr. Nenad Tomasev (DeepMind), Prof. Finale Doshi-Velez (Harvard SEAS), UK Biobank, and our partners, EyePACS, Aravind Eye Hospital and Sankara Nethralaya.