When most people think of proteins, their mind typically goes to protein-rich foods such as steak or tofu. But proteins are so much more. They’re essential to how living things operate and thrive, and studying them can help improve lives. For example, insulin treatments are life-changing for people with diabetes that are based on years of studying proteins.
There is a world of information yet to discover when it comes to proteins — from helping people get the healthcare they need to finding ways to protect plant species. Teams at Google are focused on studying proteins so we can realize Google Health’s mission to help billions of people live healthier lives.
Back in March, we published apost about a model we developed at Google that predicts protein function and a tool that allows scientists to use this model. Since then, the protein function team has accomplished more work in this space. We chatted with software engineer Max Bileschi to find out more about studying proteins and the work Google is doing.
Can you give us a quick crash course in proteins?
Proteins dictate so much of what happens in and around us, like how we and other organisms function.
Two things determine what a protein does: its chemical formula and its environment. For example, we know that human hemoglobin, a protein inside your blood, carries oxygen to your organs. We also know that if there are particular tiny changes to the chemical formula of hemoglobin in your body, it can trigger sickle cell anemia. Further, we know that blood behaves differently at different temperatures because proteins behave differently at higher temperatures.
So why did a team at Google start studying proteins?
We have the opportunity to look at how machine learning can help various scientific fields. Proteins are an obvious choice because of the amazing breadth of functions they have in our bodies and in the world. There is an enormous amount of public data, and while individual researchers have done excellent work studying specific proteins, we know that we’ve just scratched the surface of fully understanding the protein universe. It’s highly aligned to Google’s mission of organizing information and making it accessible and useful.
This sounds exciting! Tell us more about the use of machine learning in identifying what proteins do and how it improves upon the status quo.
Only around 1% of proteins have been studied in a laboratory setting. We want to see how machine learning can help us learn about the other 99%.
It’s difficult work. There are at least a billion proteins in the world, and they’ve evolved throughout history and have been shaped by the same forces of natural selection we normally think of as acting on DNA. It’s useful to understand this evolutionary relatedness among proteins. The presence of a similar protein in two or more distantly related organisms (say humans and zebrafish) can be indicative that it’s useful for survival. Proteins that are closely related can have similar functions but with small differences, like encouraging the same chemical reaction but doing so at different temperatures. Sometimes it’s easy to determine that two proteins are closely related, but other times it's difficult. This was the first problem in protein function annotation that we tackled with machine learning.
Machine learning helps best when it truly helps, not replaces, current techniques. For example, we demonstrated that about 300 previously-uncharacterized proteins are related to “phage capsid” proteins. These capsid proteins can help us deliver medicines to the cells that really need them. We worked with a trusted protein database, Pfam, to confirm our hypothesis, and now these proteins are listed as being related to phage capsid proteins — for all the public to see — including researchers.
Back up a bit. Can you explain what the protein family database Pfam is? How has your team contributed to this database?
A community of scientists built a number of tools and databases, over decades, to help classify what each different protein does. Pfam is one of the most-used databases, and it classifies proteins into about 20,000 types of proteins.
This work of classifying proteins requires both computer models and experts (called curators) to validate and improve the computer models.
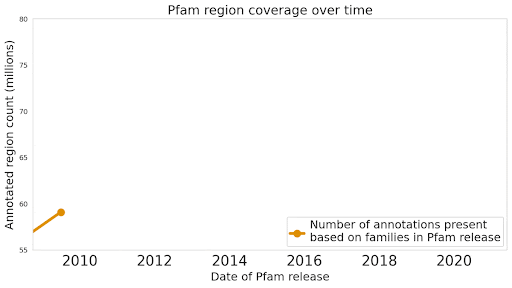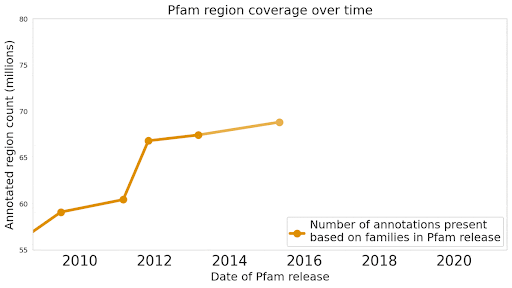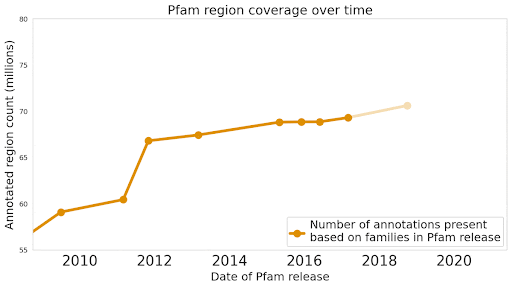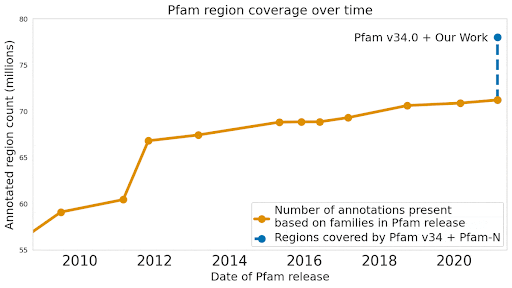
We used machine learning to add classifications for human proteins that previously lacked Pfam classifications — helping grow the database and adding several years of progress.
Since the publication of your paper ‘Using deep learning to annotate the protein universe’ in June, what has your team been up to?
We’re focused on identifying more proteins and sharing that knowledge with the science and research community. At the end of July, we released more data jointly with Pfam. And this month we’re making Pfam data and Mgnify data, another database that catalogs microbiome data, available on Google Cloud Platform so more people can have access to it. Later this year, we’ll launch an initiative with UniProt, a prominent database in our field, to use language models to name uncharacterized proteins in UniProt. We’re excited about the progress we’re making and how sharing this data can help solve challenging problems.