 Learn more about managing your stress with Fitbit and Google Pixel Watch.
Learn more about managing your stress with Fitbit and Google Pixel Watch.
How Fitbit can help you measure stress — and use it to your advantage
 Learn more about managing your stress with Fitbit and Google Pixel Watch.
Learn more about managing your stress with Fitbit and Google Pixel Watch.
 Learn more about managing your stress with Fitbit and Google Pixel Watch.
Learn more about managing your stress with Fitbit and Google Pixel Watch.
 Discover how companies are using AI technologies to fuel innovation, create new customer experiences and develop better tools for employees.
Discover how companies are using AI technologies to fuel innovation, create new customer experiences and develop better tools for employees.
Lung cancer is the leading cause of cancer-related deaths globally with 1.8 million deaths reported in 2020. Late diagnosis dramatically reduces the chances of survival. Lung cancer screening via computed tomography (CT), which provides a detailed 3D image of the lungs, has been shown to reduce mortality in high-risk populations by at least 20% by detecting potential signs of cancers earlier. In the US, screening involves annual scans, with some countries or cases recommending more or less frequent scans.
The United States Preventive Services Task Force recently expanded lung cancer screening recommendations by roughly 80%, which is expected to increase screening access for women and racial and ethnic minority groups. However, false positives (i.e., incorrectly reporting a potential cancer in a cancer-free patient) can cause anxiety and lead to unnecessary procedures for patients while increasing costs for the healthcare system. Moreover, efficiency in screening a large number of individuals can be challenging depending on healthcare infrastructure and radiologist availability.
At Google we have previously developed machine learning (ML) models for lung cancer detection, and have evaluated their ability to automatically detect and classify regions that show signs of potential cancer. Performance has been shown to be comparable to that of specialists in detecting possible cancer. While they have achieved high performance, effectively communicating findings in realistic environments is necessary to realize their full potential.
To that end, in “Assistive AI in Lung Cancer Screening: A Retrospective Multinational Study in the US and Japan”, published in Radiology AI, we investigate how ML models can effectively communicate findings to radiologists. We also introduce a generalizable user-centric interface to help radiologists leverage such models for lung cancer screening. The system takes CT imaging as input and outputs a cancer suspicion rating using four categories (no suspicion, probably benign, suspicious, highly suspicious) along with the corresponding regions of interest. We evaluate the system’s utility in improving clinician performance through randomized reader studies in both the US and Japan, using the local cancer scoring systems (Lung-RADSs V1.1 and Sendai Score) and image viewers that mimic realistic settings. We found that reader specificity increases with model assistance in both reader studies. To accelerate progress in conducting similar studies with ML models, we have open-sourced code to process CT images and generate images compatible with the picture archiving and communication system (PACS) used by radiologists.
Integrating ML models into radiologist workflows involves understanding the nuances and goals of their tasks to meaningfully support them. In the case of lung cancer screening, hospitals follow various country-specific guidelines that are regularly updated. For example, in the US, Lung-RADs V1.1 assigns an alpha-numeric score to indicate the lung cancer risk and follow-up recommendations. When assessing patients, radiologists load the CT in their workstation to read the case, find lung nodules or lesions, and apply set guidelines to determine follow-up decisions.
Our first step was to improve the previously developed ML models through additional training data and architectural improvements, including self-attention. Then, instead of targeting specific guidelines, we experimented with a complementary way of communicating AI results independent of guidelines or their particular versions. Specifically, the system output offers a suspicion rating and localization (regions of interest) for the user to consider in conjunction with their own specific guidelines. The interface produces output images directly associated with the CT study, requiring no changes to the user’s workstation. The radiologist only needs to review a small set of additional images. There is no other change to their system or interaction with the system.
The assistive lung cancer screening system comprises 13 models and has a high-level architecture similar to the end-to-end system used in prior work. The models coordinate with each other to first segment the lungs, obtain an overall assessment, locate three suspicious regions, then use the information to assign a suspicion rating to each region. The system was deployed on Google Cloud using a Google Kubernetes Engine (GKE) that pulled the images, ran the ML models, and provided results. This allows scalability and directly connects to servers where the images are stored in DICOM stores.
To evaluate the system’s utility in improving clinical performance, we conducted two reader studies (i.e., experiments designed to assess clinical performance comparing expert performance with and without the aid of a technology) with 12 radiologists using pre-existing, de-identified CT scans. We presented 627 challenging cases to 6 US-based and 6 Japan-based radiologists. In the experimental setup, readers were divided into two groups that read each case twice, with and without assistance from the model. Readers were asked to apply scoring guidelines they typically use in their clinical practice and report their overall suspicion of cancer for each case. We then compared the results of the reader’s responses to measure the impact of the model on their workflow and decisions. The score and suspicion level were judged against the actual cancer outcomes of the individuals to measure sensitivity, specificity, and area under the ROC curve (AUC) values. These were compared with and without assistance.
The ability to conduct these studies using the same interface highlights its generalizability to completely different cancer scoring systems, and the generalization of the model and assistive capability to different patient populations. Our study results demonstrated that when radiologists used the system in their clinical evaluation, they had an increased ability to correctly identify lung images without actionable lung cancer findings (i.e., specificity) by an absolute 5–7% compared to when they didn’t use the assistive system. This potentially means that for every 15–20 patients screened, one may be able to avoid unnecessary follow-up procedures, thus reducing their anxiety and the burden on the health care system. This can, in turn, help improve the sustainability of lung cancer screening programs, particularly as more people become eligible for screening.
The system results demonstrate the potential for fewer follow-up visits, reduced anxiety, as well lower overall costs for lung cancer screening. In an effort to translate this research into real-world clinical impact, we are working with: DeepHealth, a leading AI-powered health informatics provider; and Apollo Radiology International a leading provider of Radiology services in India to explore paths for incorporating this system into future products. In addition, we are looking to help other researchers studying how best to integrate ML model results into clinical workflows by open sourcing code used for the reader study and incorporating the insights described in this blog. We hope that this will help accelerate medical imaging researchers looking to conduct reader studies for their AI models, and catalyze translational research in the field.
Key contributors to this project include Corbin Cunningham, Zaid Nabulsi, Ryan Najafi, Jie Yang, Charles Lau, Joseph R. Ledsam, Wenxing Ye, Diego Ardila, Scott M. McKinney, Rory Pilgrim, Hiroaki Saito, Yasuteru Shimamura, Mozziyar Etemadi, Yun Liu, David Melnick, Sunny Jansen, Nadia Harhen, David P. Nadich, Mikhail Fomitchev, Ziyad Helali, Shabir Adeel, Greg S. Corrado, Lily Peng, Daniel Tse, Shravya Shetty, Shruthi Prabhakara, Neeral Beladia, and Krish Eswaran. Thanks to Arnav Agharwal and Andrew Sellergren for their open sourcing support and Vivek Natarajan and Michael D. Howell for their feedback. Sincere appreciation also goes to the radiologists who enabled this work with their image interpretation and annotation efforts throughout the study, and Jonny Wong and Carli Sampson for coordinating the reader studies.
 As medical AI evolves, we unveil tools and resources to help identify and mitigate biases that could negatively impact health outcomes.
As medical AI evolves, we unveil tools and resources to help identify and mitigate biases that could negatively impact health outcomes.
 We’re improving access to easy-to-understand, relatable, evidence-based information for people to better navigate their health.
We’re improving access to easy-to-understand, relatable, evidence-based information for people to better navigate their health.
Health datasets play a crucial role in research and medical education, but it can be challenging to create a dataset that represents the real world. For example, dermatology conditions are diverse in their appearance and severity and manifest differently across skin tones. Yet, existing dermatology image datasets often lack representation of everyday conditions (like rashes, allergies and infections) and skew towards lighter skin tones. Furthermore, race and ethnicity information is frequently missing, hindering our ability to assess disparities or create solutions.
To address these limitations, we are releasing the Skin Condition Image Network (SCIN) dataset in collaboration with physicians at Stanford Medicine. We designed SCIN to reflect the broad range of concerns that people search for online, supplementing the types of conditions typically found in clinical datasets. It contains images across various skin tones and body parts, helping to ensure that future AI tools work effectively for all. We've made the SCIN dataset freely available as an open-access resource for researchers, educators, and developers, and have taken careful steps to protect contributor privacy.
 |
| Example set of images and metadata from the SCIN dataset. |
The SCIN dataset currently contains over 10,000 images of skin, nail, or hair conditions, directly contributed by individuals experiencing them. All contributions were made voluntarily with informed consent by individuals in the US, under an institutional-review board approved study. To provide context for retrospective dermatologist labeling, contributors were asked to take images both close-up and from slightly further away. They were given the option to self-report demographic information and tanning propensity (self-reported Fitzpatrick Skin Type, i.e., sFST), and to describe the texture, duration and symptoms related to their concern.
One to three dermatologists labeled each contribution with up to five dermatology conditions, along with a confidence score for each label. The SCIN dataset contains these individual labels, as well as an aggregated and weighted differential diagnosis derived from them that could be useful for model testing or training. These labels were assigned retrospectively and are not equivalent to a clinical diagnosis, but they allow us to compare the distribution of dermatology conditions in the SCIN dataset with existing datasets.
 |
| The SCIN dataset contains largely allergic, inflammatory and infectious conditions while datasets from clinical sources focus on benign and malignant neoplasms. |
While many existing dermatology datasets focus on malignant and benign tumors and are intended to assist with skin cancer diagnosis, the SCIN dataset consists largely of common allergic, inflammatory, and infectious conditions. The majority of images in the SCIN dataset show early-stage concerns — more than half arose less than a week before the photo, and 30% arose less than a day before the image was taken. Conditions within this time window are seldom seen within the health system and therefore are underrepresented in existing dermatology datasets.
We also obtained dermatologist estimates of Fitzpatrick Skin Type (estimated FST or eFST) and layperson labeler estimates of Monk Skin Tone (eMST) for the images. This allowed comparison of the skin condition and skin type distributions to those in existing dermatology datasets. Although we did not selectively target any skin types or skin tones, the SCIN dataset has a balanced Fitzpatrick skin type distribution (with more of Types 3, 4, 5, and 6) compared to similar datasets from clinical sources.
 |
| Self-reported and dermatologist-estimated Fitzpatrick Skin Type distribution in the SCIN dataset compared with existing un-enriched dermatology datasets (Fitzpatrick17k, PH², SKINL2, and PAD-UFES-20). |
The Fitzpatrick Skin Type scale was originally developed as a photo-typing scale to measure the response of skin types to UV radiation, and it is widely used in dermatology research. The Monk Skin Tone scale is a newer 10-shade scale that measures skin tone rather than skin phototype, capturing more nuanced differences between the darker skin tones. While neither scale was intended for retrospective estimation using images, the inclusion of these labels is intended to enable future research into skin type and tone representation in dermatology. For example, the SCIN dataset provides an initial benchmark for the distribution of these skin types and tones in the US population.
The SCIN dataset has a high representation of women and younger individuals, likely reflecting a combination of factors. These could include differences in skin condition incidence, propensity to seek health information online, and variations in willingness to contribute to research across demographics.
To create the SCIN dataset, we used a novel crowdsourcing method, which we describe in the accompanying research paper co-authored with investigators at Stanford Medicine. This approach empowers individuals to play an active role in healthcare research. It allows us to reach people at earlier stages of their health concerns, potentially before they seek formal care. Crucially, this method uses advertisements on web search result pages — the starting point for many people’s health journey — to connect with participants.
Our results demonstrate that crowdsourcing can yield a high-quality dataset with a low spam rate. Over 97.5% of contributions were genuine images of skin conditions. After performing further filtering steps to exclude images that were out of scope for the SCIN dataset and to remove duplicates, we were able to release nearly 90% of the contributions received over the 8-month study period. Most images were sharp and well-exposed. Approximately half of the contributions include self-reported demographics, and 80% contain self-reported information relating to the skin condition, such as texture, duration, or other symptoms. We found that dermatologists’ ability to retrospectively assign a differential diagnosis depended more on the availability of self-reported information than on image quality.
 |
| Dermatologist confidence in their labels (scale from 1-5) depended on the availability of self-reported demographic and symptom information. |
While perfect image de-identification can never be guaranteed, protecting the privacy of individuals who contributed their images was a top priority when creating the SCIN dataset. Through informed consent, contributors were made aware of potential re-identification risks and advised to avoid uploading images with identifying features. Post-submission privacy protection measures included manual redaction or cropping to exclude potentially identifying areas, reverse image searches to exclude publicly available copies and metadata removal or aggregation. The SCIN Data Use License prohibits attempts to re-identify contributors.
We hope the SCIN dataset will be a helpful resource for those working to advance inclusive dermatology research, education, and AI tool development. By demonstrating an alternative to traditional dataset creation methods, SCIN paves the way for more representative datasets in areas where self-reported data or retrospective labeling is feasible.
We are grateful to all our co-authors Abbi Ward, Jimmy Li, Julie Wang, Sriram Lakshminarasimhan, Ashley Carrick, Bilson Campana, Jay Hartford, Pradeep Kumar S, Tiya Tiyasirisokchai, Sunny Virmani, Renee Wong, Yossi Matias, Greg S. Corrado, Dale R. Webster, Dawn Siegel (Stanford Medicine), Steven Lin (Stanford Medicine), Justin Ko (Stanford Medicine), Alan Karthikesalingam and Christopher Semturs. We also thank Yetunde Ibitoye, Sami Lachgar, Lisa Lehmann, Javier Perez, Margaret Ann Smith (Stanford Medicine), Rachelle Sico, Amit Talreja, Annisah Um’rani and Wayne Westerlind for their essential contributions to this work. Finally, we are grateful to Heather Cole-Lewis, Naama Hammel, Ivor Horn, Michael Howell, Yun Liu, and Eric Teasley for their insightful comments on the study design and manuscript.
 At The Check Up, we’re sharing progress on our generative AI efforts in health, including product updates and new research.
At The Check Up, we’re sharing progress on our generative AI efforts in health, including product updates and new research.
 At The Check Up, we shared more about the transformative potential of AI in health and our latest research, partnership and product updates.
At The Check Up, we shared more about the transformative potential of AI in health and our latest research, partnership and product updates.
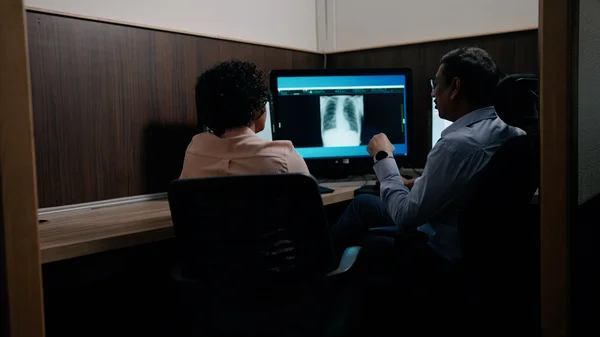 We’re collaborating with Apollo Radiology International to bring AI-powered screening for tuberculosis, lung and breast cancer to India.
We’re collaborating with Apollo Radiology International to bring AI-powered screening for tuberculosis, lung and breast cancer to India.
Health equity is a major societal concern worldwide with disparities having many causes. These sources include limitations in access to healthcare, differences in clinical treatment, and even fundamental differences in the diagnostic technology. In dermatology for example, skin cancer outcomes are worse for populations such as minorities, those with lower socioeconomic status, or individuals with limited healthcare access. While there is great promise in recent advances in machine learning (ML) and artificial intelligence (AI) to help improve healthcare, this transition from research to bedside must be accompanied by a careful understanding of whether and how they impact health equity.
Health equity is defined by public health organizations as fairness of opportunity for everyone to be as healthy as possible. Importantly, equity may be different from equality. For example, people with greater barriers to improving their health may require more or different effort to experience this fair opportunity. Similarly, equity is not fairness as defined in the AI for healthcare literature. Whereas AI fairness often strives for equal performance of the AI technology across different patient populations, this does not center the goal of prioritizing performance with respect to pre-existing health disparities.
In “Health Equity Assessment of machine Learning performance (HEAL): a framework and dermatology AI model case study”, published in The Lancet eClinicalMedicine, we propose a methodology to quantitatively assess whether ML-based health technologies perform equitably. In other words, does the ML model perform well for those with the worst health outcomes for the condition(s) the model is meant to address? This goal anchors on the principle that health equity should prioritize and measure model performance with respect to disparate health outcomes, which may be due to a number of factors that include structural inequities (e.g., demographic, social, cultural, political, economic, environmental and geographic).
The HEAL framework proposes a 4-step process to estimate the likelihood that an ML-based health technology performs equitably:
The final step’s output is termed the HEAL metric, which quantifies how anticorrelated the ML model’s performance is with health disparities. In other words, does the model perform better with populations that have the worse health outcomes?
This 4-step process is designed to inform improvements for making ML model performance more equitable, and is meant to be iterative and re-evaluated on a regular basis. For example, the availability of health outcomes data in step (2) can inform the choice of demographic factors and brackets in step (1), and the framework can be applied again with new datasets, models and populations.
With this work, we take a step towards encouraging explicit assessment of the health equity considerations of AI technologies, and encourage prioritization of efforts during model development to reduce health inequities for subpopulations exposed to structural inequities that can precipitate disparate outcomes. We should note that the present framework does not model causal relationships and, therefore, cannot quantify the actual impact a new technology will have on reducing health outcome disparities. However, the HEAL metric may help identify opportunities for improvement, where the current performance is not prioritized with respect to pre-existing health disparities.
As an illustrative case study, we applied the framework to a dermatology model, which utilizes a convolutional neural network similar to that described in prior work. This example dermatology model was trained to classify 288 skin conditions using a development dataset of 29k cases. The input to the model consists of three photos of a skin concern along with demographic information and a brief structured medical history. The output consists of a ranked list of possible matching skin conditions.
Using the HEAL framework, we evaluated this model by assessing whether it prioritized performance with respect to pre-existing health outcomes. The model was designed to predict possible dermatologic conditions (from a list of hundreds) based on photos of a skin concern and patient metadata. Evaluation of the model is done using a top-3 agreement metric, which quantifies how often the top 3 output conditions match the most likely condition as suggested by a dermatologist panel. The HEAL metric is computed via the anticorrelation of this top-3 agreement with health outcome rankings.
We used a dataset of 5,420 teledermatology cases, enriched for diversity in age, sex and race/ethnicity, to retrospectively evaluate the model’s HEAL metric. The dataset consisted of “store-and-forward” cases from patients of 20 years or older from primary care providers in the USA and skin cancer clinics in Australia. Based on a review of the literature, we decided to explore race/ethnicity, sex and age as potential factors of inequity, and used sampling techniques to ensure that our evaluation dataset had sufficient representation of all race/ethnicity, sex and age groups. To quantify pre-existing health outcomes for each subgroup we relied on measurements from public databases endorsed by the World Health Organization, such as Years of Life Lost (YLLs) and Disability-Adjusted Life Years (DALYs; years of life lost plus years lived with disability).
However, while the model was likely to perform equitably across age groups for cancer conditions specifically, we discovered that it had room for improvement across age groups for non-cancer conditions. For example, those 70+ have the poorest health outcomes related to non-cancer skin conditions, yet the model didn't prioritize performance for this subgroup.
For holistic evaluation, the HEAL metric cannot be employed in isolation. Instead this metric should be contextualized alongside many other factors ranging from computational efficiency and data privacy to ethical values, and aspects that may influence the results (e.g., selection bias or differences in representativeness of the evaluation data across demographic groups).
As an adversarial example, the HEAL metric can be artificially improved by deliberately reducing model performance for the most advantaged subpopulation until performance for that subpopulation is worse than all others. For illustrative purposes, given subpopulations A and B where A has worse health outcomes than B, consider the choice between two models: Model 1 (M1) performs 5% better for subpopulation A than for subpopulation B. Model 2 (M2) performs 5% worse on subpopulation A than B. The HEAL metric would be higher for M1 because it prioritizes performance on a subpopulation with worse outcomes. However, M1 may have absolute performances of just 75% and 70% for subpopulations A and B respectively, while M2 has absolute performances of 75% and 80% for subpopulations A and B respectively. Choosing M1 over M2 would lead to worse overall performance for all subpopulations because some subpopulations are worse-off while no subpopulation is better-off.
Accordingly, the HEAL metric should be used alongside a Pareto condition (discussed further in the paper), which restricts model changes so that outcomes for each subpopulation are either unchanged or improved compared to the status quo, and performance does not worsen for any subpopulation.
The HEAL framework, in its current form, assesses the likelihood that an ML-based model prioritizes performance for subpopulations with respect to pre-existing health disparities for specific subpopulations. This differs from the goal of understanding whether ML will reduce disparities in outcomes across subpopulations in reality. Specifically, modeling improvements in outcomes requires a causal understanding of steps in the care journey that happen both before and after use of any given model. Future research is needed to address this gap.
The HEAL framework enables a quantitative assessment of the likelihood that health AI technologies prioritize performance with respect to health disparities. The case study demonstrates how to apply the framework in the dermatological domain, indicating a high likelihood that model performance is prioritized with respect to health disparities across sex and race/ethnicity, but also revealing the potential for improvements for non-cancer conditions across age. The case study also illustrates limitations in the ability to apply all recommended aspects of the framework (e.g., mapping societal context, availability of data), thus highlighting the complexity of health equity considerations of ML-based tools.
This work is a proposed approach to address a grand challenge for AI and health equity, and may provide a useful evaluation framework not only during model development, but during pre-implementation and real-world monitoring stages, e.g., in the form of health equity dashboards. We hold that the strength of the HEAL framework is in its future application to various AI tools and use cases and its refinement in the process. Finally, we acknowledge that a successful approach towards understanding the impact of AI technologies on health equity needs to be more than a set of metrics. It will require a set of goals agreed upon by a community that represents those who will be most impacted by a model.
The research described here is joint work across many teams at Google. We are grateful to all our co-authors: Terry Spitz, Malcolm Pyles, Heather Cole-Lewis, Ellery Wulczyn, Stephen R. Pfohl, Donald Martin, Jr., Ronnachai Jaroensri, Geoff Keeling, Yuan Liu, Stephanie Farquhar, Qinghan Xue, Jenna Lester, Cían Hughes, Patricia Strachan, Fraser Tan, Peggy Bui, Craig H. Mermel, Lily H. Peng, Yossi Matias, Greg S. Corrado, Dale R. Webster, Sunny Virmani, Christopher Semturs, Yun Liu, and Po-Hsuan Cameron Chen. We also thank Lauren Winer, Sami Lachgar, Ting-An Lin, Aaron Loh, Morgan Du, Jenny Rizk, Renee Wong, Ashley Carrick, Preeti Singh, Annisah Um'rani, Jessica Schrouff, Alexander Brown, and Anna Iurchenko for their support of this project.